Cytogenetics is a branch of genetics that includes the study of chromosome structure, function, properties, behavior during the cell division (mitosis and meiosis) and its involvement in a disease condition.
A chromosome is a unit of inheritance made up of the interaction between DNA and proteins. The long sequence of DNA codes for a protein is known as a gene. Genes are located on the chromosome.
Read our beautiful article on DNA: DNA story: The structure and function of DNA
A diploid set of chromosomes consists of 23 pairs (=46 chromosomes) while the haploid set consists of 23 chromosomes. One set from the mother and one set from the father are inherited to the fetus. And therefore a fetus carries 46 chromosomes in the genome.
22 pairs of autosomal and a pair of sex chromosomes are present in every cell. XX in females and XY in a male are present as sex chromosomes.
The chromosome is a complex network of DNA and proteins. The process called DNA packaging helps DNA to locate on a chromosome. It has 4 different chromatids, carries telomeres at the ends. The central part of the chromosome is known as the centromere.
Based on the location of the centromere the chromosomes are classified into metacentric, submetacentric and acrocentric chromosomes, etc.
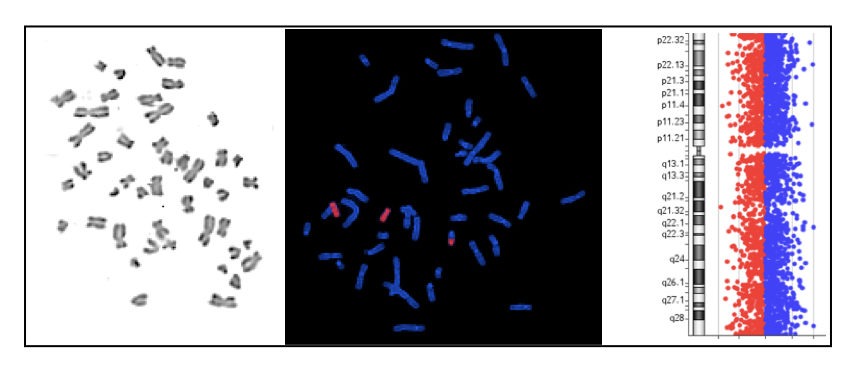
In the present article, the major focus will be on the techniques used in cytogenetic investigations.
Key Topics:
History of Cytogenetics:
The story of cytogenetics was started unknowingly in the mid 18’s when Karl Wilhelm von Nageli, a Swiss botanist, first observed the plant cell in the year 1842.
He observed a thread-like structure in the nuclei of the plant cell and named it “transitory cytoplasts”.
In 1888, Waldeyer named it “chromosome”
In 1956, the diploid numbers of chromosomes were established in humans.
Several milestone discoveries in cytogenetics are summarised below,
1931: B. McClintock explained the transposition of DNA sequences by analyzing the chromosomal structure of maize chromosomes.
1950: Colchicine was used to arrest chromosomes at metaphase.
1956: Moorehead and coworkers developed a method called “Peripheral blood leukocyte culture” for karyotyping. Later on, the method becomes popular and adopted by cytogenetics worldwide.
1956: Tjio JH, Levan A established human chromosomes as 46.
1959: Lejeune discovered trisomy 21, down syndrome.
1959: Ford and coworker observed the presence of a single X chromosome (45, X), the Turner syndrome.
1959: Jacobs and Strong observed the presence of one extra X chromosome, 46, XXY in humans (Klinefelter syndrome).
1960: Peter Nowell and David Hungerford observed translocation between chromosomes 9 and 22 and named it a Philadelphia chromosome.
1963: Lejeune and coworker examined the deletion of a short arm of chromosome 5 which results in cry-do-chat syndrome.
1970: Caspersson and coworkers discovered the first chromosomal banding technique, the Q-banding or quinacrine banding.
1971: Maximo Drets and Margery Shaw explained Giemsa staining and Giemsa banding methods.
Read some interesting articles:
- Gene Therapy: Types, Vectors [Viral and Non-Viral], Process, Applications and Limitations
- Transposons: A Jumping Entity and a Foe with Benefits
Introduction to cytogenetics:
“Cytogenetics is a branch of science that deals with the study of chromosomes, chromosomal alterations and its relation in the disease development.”
In simple words, “The study of chromosomes is called cytogenetics.”
Through the techniques of cytogenetics such as karyotyping, FISH and whole chromosome microarray we can study,
- The structural properties of a chromosome (deletion, duplication, translocation or alteration).
- The functional properties of a chromosome (loss or gain of function)
- Numerical chromosomal abnormalities (trisomy, tetrasomy or monosomy).
- The behaviour of the chromosomes during the mitosis or somatic cell division (growth and development).
- The behaviour of the chromosomes meiosis or germ cell division (reproduction).
Initially, the technique was developed to find out chromosomal alterations such as ploidies and aneuploidies in plants.
However, nowadays it is used for the characterization, identification and detection of human genetic diseases.
State-of-the-art lab setups and continuous improvements in cytogenetic techniques benefit prenatal diagnosis a lot. Now it is possible to screen numerical chromosomal abnormalities like Down syndrome, Edward syndrome and Klinefelter syndrome during the prenatal stage.
And hence significant improvement is observed in the management of those disorders.
In the cytogenetic analysis “cell” is one of the major constituents. Some of the common cell types used for the cytogenetic analysis are listed below,
| Tissue | Cell type used for cytogenetic analysis | Detection |
| Peripheral blood | Lymphocytes | All types of numerical and some routine structural abnormalities analysis |
| Embryo | Blastomere cells | Preimplantation genetic analysis of embryo |
| Bone marrow | WBC | Leukemia |
| Chorionic Villi | Trophoblast | First-trimester prenatal diagnosis |
| Amniotic fluid | Amniocytes | Second-trimester prenatal diagnosis |
One of the first techniques developed for chromosomal analysis was karyotyping.
However, more advanced and accurate molecular cytogenetic techniques such as FISH and DNA microarray are now available.
The method of cytogenetics is divided into two broader categories:
- Constitutional cytogenetics
- Cancer cytogenetics
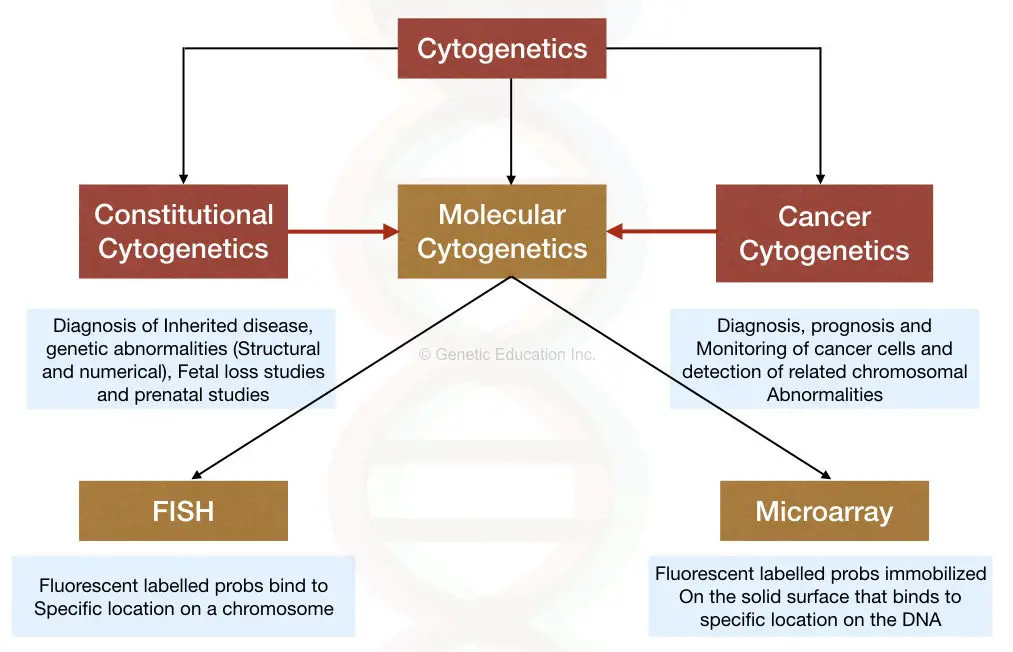
Constitutional cytogenetics:
The constitutional cytogenetic includes the study of different genetic conditions (except cancer).
Constitutional cytogenetics helps in the diagnosis of structural and numerical chromosomal abnormalities, inherited genetic abnormalities, fetal loss and pregnancy-related issues.
Indications:
Autism, hypotonia, down syndrome, Patau syndrome, Edward syndrome, birth defects, malformations, ambiguous genitalia, short stature, growth failure, developmental delay, cry-du-chat syndrome, Klinefelter syndrome.
Infertility causes such as azoospermia, hypogonadism and oligospermia can also be indicated using constitutional cytogenetics.
Also, ovarian failure, the product of conception and spontaneous abortion can also be indicated.
Cancer Cytogenetics:
This field involves detection, prognosis and monitoring of cancer cells.
Cancer cytogenetics detects a type of cancer in which chromosomal abnormalities are involved.
Indications:
ALL (Acute Lymphocytic Leukemia), CLL (Chronic Lymphocytic Leukemia), NHL (Non-Hodgkin lymphoma) and PCN like lymphoid and CML (Chronic Myeloid Leukemia), AML (Acute Myeloid Leukemia), MPN (Myeloproliferative Neoplasms) and MDS (myelodysplastic syndromes) like myeloid cancer can be indicated using the cancer cytogenetics.
Importance of cytogenetics:
Cytogenetic techniques are a very important tool for detection and indication of chromosomal abnormalities, it used for the study of,
- Aneuploidy and polyploidy
- Gametogenesis
- Nondisjunction and sister chromatid exchange
- Genetic imprinting and uniparental disomy s
- Identification, characterisation and nomenclature of chromosomes
- Sex chromosomal abnormalities
- Cell cycle and replication analysis
- Drug discovery
Cytogenetic techniques:
The cytogenetic techniques are of two types, conventional cytogenetic techniques such as Karyotyping and molecular cytogenetics, FISH and microarray.
Peripheral blood leukocyte culture:
The PBLC is also known as standard karyotyping in which analysis of metaphase chromosome is done using chromosome harvesting and staining.
The major steps of PBLC are:
- Collection of sample
- Cell culture
- Chromosome harvesting
- Chromosome staining and banding
- Result analysis
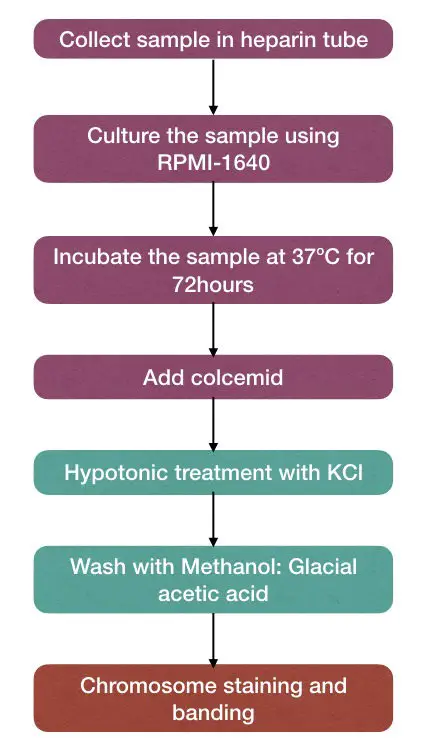
Sample collection:
Blood samples for karyotyping are collected into the heparin tubes.
The sample type for the PBLC is blood.
The aim of Karyotyping is to arrest the cells at the metaphase stage so that each chromosome can be distinguished.
For that, the collected blood cells are cultured immediately.
Cell culture:
The sample must be cultured within 24 hours of collection.
For routine laboratory karyotyping RPMI-1640, a complete media is used for culturing the cells, All the culturing procedures are performed under strict aseptic conditions (The chance of contamination is very high in cell culture experiments).
Now the cells are incubated at 37°C for 72 hours, once the cells are cultured.
After the completion of incubation, the cell division process stopped using the colcemid.
The culturing process is completed here. Now the cell culture is processed for harvesting.
Chromosome harvesting:
The hypotonic treatment is given to the cultured cells so that the cell swells and chromosomes separate properly.
The hypotonic treatment is given for 30 sec (the time varies from lab to lab).
Immediately after that, the combination of methanol: glacial acetic acid is added and the cell suspension is centrifuged at 2500rpm for 20 minutes.
For harvesting, the combination of methanol: glacial acetic acid (known as a fixative) is taken and the cultured cells are washed until the clear cell suspension is obtained.
After the completion of harvesting, the staining procedure is a must-needed step for confirming our results.
Chromosome staining and banding:
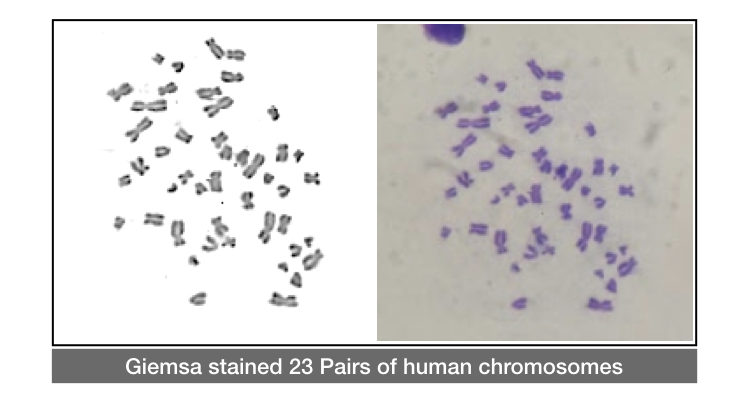
Now the cells are stained with Giemsa stain to observe under the microscope.
The stained chromosomes are looks dark blue under the microscope, see my experiment results:
For the detection of structural or numerical abnormalities, chromosomal banding is performed.
Some of the common banding techniques:
- Giemsa banding (G banding)
- Q banding
- C banding
- NOR banding
Result and interpretation:
A standard karyogram is prepared for analyzing the results. For preparing the karyogram, each chromosome is arranged properly based on the location of centromeres. See the image below,

Molecular cytogenetics:
The Karyotyping method is one of the best methods for screening numerical chromosomal abnormalities although, specialized expertise and vast experience are required to interpret results.
However, minor copy number variations such as minor duplications, deletions or translocations cannot be identified using the standard karyotyping method.
FISH and microarray are molecular cytogenetic techniques that help to overcome this problem.
The molecular cytogenetic techniques can identify even a smaller deletion, duplication or other copy number variations.
Read this article: Short Tandem Repeats (STRs): A Secret of Every DNA Test
FISH (Fluorescent In Situ Hybridization):
The Fluorescent In Situ hybridization technique is used to map or identify specific DNA sequences on a chromosome.
Here in FISH, the fluorescently labeled DNA probes are directly used to hybridize on the specific location on a chromosome.
The fluoro-labeled DNA probes are specific to some DNA sequence ( which we want to study) or specific to some gene.
Once the probes are hybridized on chromosomes that gene sequence can be visualized under the fluorescent microscope.
By doing this we can actually visualize the location of a particular gene on a chromosome.
The hybridization is performed directly on the chromosome slide and observed under the microscope.
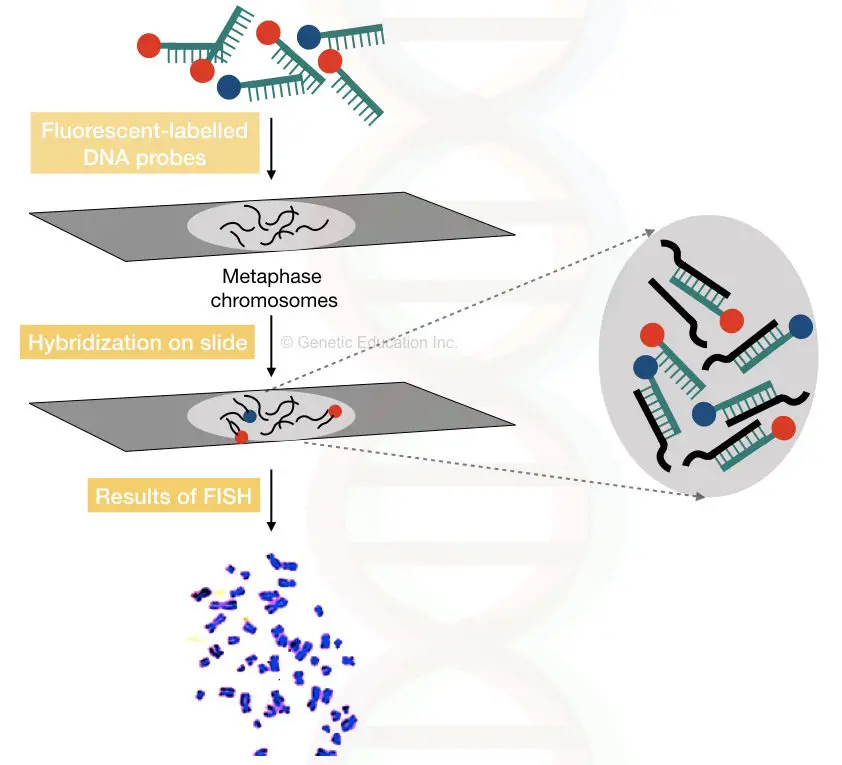
Nowadays, so many ready-to-use probes for different types of markers are available commercially. The advancement in native FISH technology gives tremendous power to interpret results.
One of the most common improvisations is multicolor FISH or M-FISH.
The M-FISH is also known as spectral karyotyping or multiplex-FISH in which different fluorescent color probes for different chromosomes are used for hybridization.
For instance, suppose a green fluorescent probe can only hybridize the sequences of chromosome number 2 while the red-colored probe can only bind to the sequences of chromosome 1.
For spectral karyotyping 24 different colored fluorescent probes are used for 24 different chromosomes.
The spectral karyotyping method is commonly applicable for the identifications of translocations.
How FISH is beneficial over conventional karyotyping?
Let us take an example of two different syndromes: Prader-Willi syndrome and Angelman syndrome.
Both have the same deletions on chromosome number 15 at the same location, however, the phenotypes are different.
By doing PCR or conventional karyotyping one can only observe the deletion but not more than that.
After the analysis of FISH results, scientists concluded that in the Prader-Willi syndrome chromosome 15 deletion was inherited from the father while in the Angelman chromosome 15 deletion was inherited from the mother.
In a FISH one can analyze limited numbers of chromosome areas of a specific gene sequence. Further, the numbers of a probe used in a single experiment are limited, therefore a much automated-robust and advanced detection system is required.
The comparative genomic hybridization and DNA microarray fulfill all these requirements.
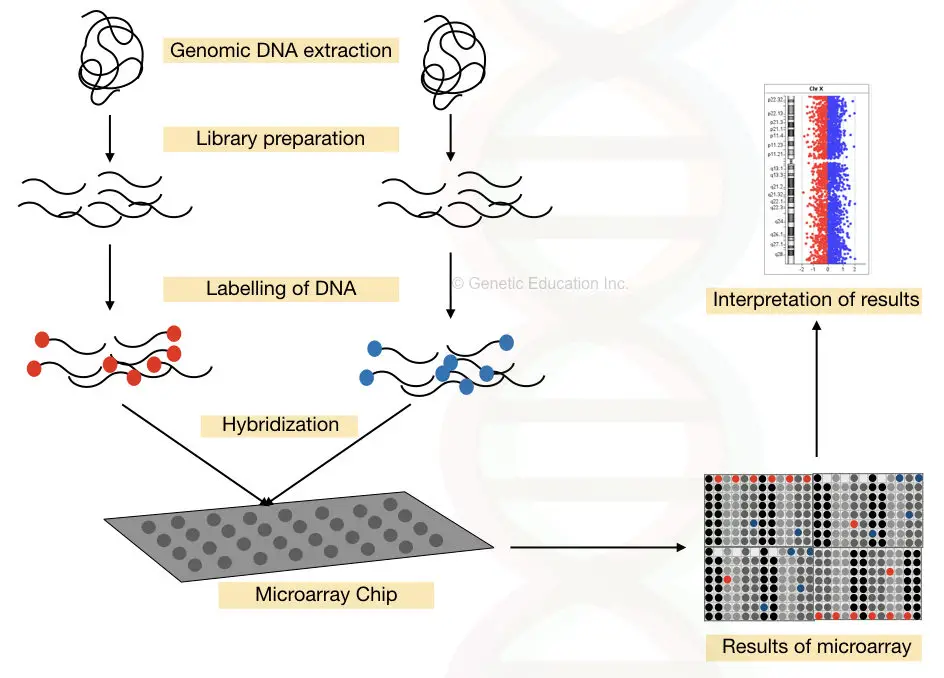
Comparative Genomic Hybridization:
More regions of chromosomes can be analyzed using the CGH, comparative genomic hybridization method.
The DNA is extracted from the control as well as a patient and a library of DNA fragments is created.
The DNA library of a patient is labeled with red fluorescent dye while the DNA library of control is labeled with the green fluorescent dye.
The library of both the labeled DNA is used to hybridize with the normal chromosomes.
The higher fluorescent intensity of the green dye-labeled DNA indicates the addition of several other sequences, while the higher intensity of the red dye-labeled indicates the loss of the sequences (deletion).
The aim of the CGH is to detect the gain or loss of copy number variations.
The CGH method has 10 to 15 fold more resolution than conventional karyotyping or FISH.
However, the comparative genomic hybridization method is laborious, time-consuming and costly, also, one has to culture chromosomes to do the CGH.
DNA microarray:
Synonyms: DNA chips, DNA array, gene array, biochips. Gene chips.
The DNA microarray technology developed in the mid-’90s and the idea was given from the Southern blot hybridization method.
In the DNA microarray method, instead of cultured chromosomes only genomic DNA or mRNA is required (depending upon the array).
The DNA microarrays are of two types:
- For calculating the gene expression: cDNA based microarray
- For identification of copy number variations: DNA microarray
In the gene expression microarray, total mRNA is extracted and purified for the array hybridization.
The cDNA is synthesized from the mRNA using the reverse transcription mechanism.
The library of cDNA is prepared for the hybridization process.
Millions of oligo probes are immobilized on the solid surface, a specific oligo binds to its complementary sequence on the cDNA.
Once it binds it generates a signal which is detected by the detector.
The unbound oligos and cDNA are wash off by different washing steps therefore, no unnecessary signals are generated.
In the DNA microarray,
The DNA fragment library is prepared from the total genomic DNA in the first step.
Here, thousands of fluorescently labeled probes for different copy number variations are immobilized on the glass slide.
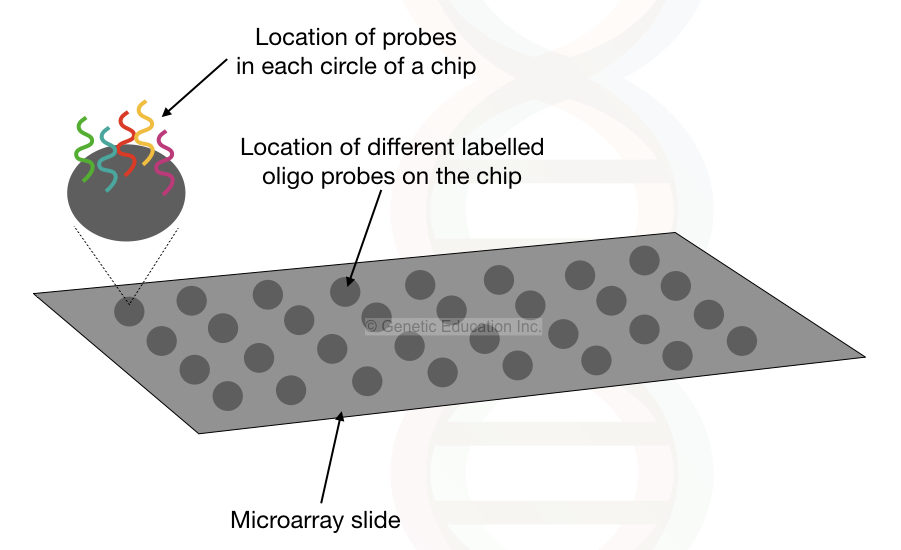
The labeled probes are attached to the glass or silicon surface, each probe represents a single DNA sequence to be screened.
Once the DNA sample is applied on the surface of the glass slide to hybridize with the probes if any copy number variation is present, the specific oligo probe binds to DNA and emits more fluorescent.
The signal from the microarray slides is recorded by the detector for analysis of results.
The method has 1000 to 2000 fold more resolution than the FISH.
Thousands of chromosomal aberrations can be detected in a single run of a microarray.
The complete process of DNA microarray is shown in the figure below,
Information:
The SNP DNA microarray is a special type of microarray method designed to screen millions of pathogenic SNPs present in the human genome.
Applications of cytogenetics:
Broadly, the applications of cytogenetics (karyotyping, FISH and DNA microarray) are listed below,
- Plan genomic research
- Crop improvements
- Diagnosis of inherited disease
- Diagnosis of infectious disease
- Transcriptome studies
- Genotyping and SNP detection
- Identification of microbes
- Copy number variations detections
- Population study through genomic loss or gain study
- Cancer research
- Detection of tissue-specific gene expression
- Toxicogenomics, drug discovery and genomic medicines
Drawbacks:
The methods are extremely tedious and laborious. A lot of expertise and experience are required to interpret the results.
The chance of experiment failure and false-positive results are very high.
The molecular cytogenetic methods are expensive and the shelf life of the oligo chip is not so good.
Other applications of cytogenetics:
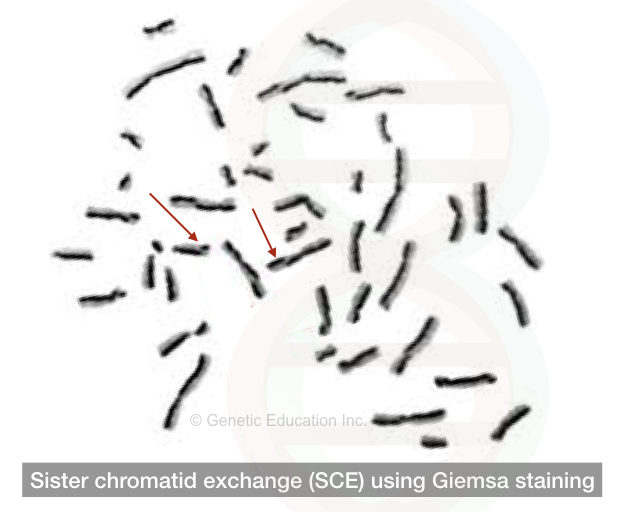
Sister chromatid exchange:
The mechanism of studying the exchange of genetic material between the two identical sister chromatids is called SCE (sister chromatid exchange).
Unwantedly exchange of genetic material between two identical sister chromatids results in genetic abnormalities in an organism that can be screened using the sister chromatid exchange through karyotyping.
Using the Giemsa staining and bromodeoxyuridine base analogous two different sister chromatids can be studied.
The results of the SCE are seen in the figure below,
Replication stage:
Early replicating and late replicating chromosomes can also be distinguished using the cytogenetic methods.
Read more:
Conclusion:
The diagnostic utility of cytogenetic techniques especially, karyotyping is unmatched. The technique is practiced for the identifications of chromosomal abnormalities for a long.
However, the process of karyotyping is difficult. The protocol for cell culturing and harvesting vary from lab to lab. Therefore, specialized training and work expertise are required to perform it.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.



Very informative ancd concise
Thank you