“DNA packaging is a process involving DNA and protein, for arranging DNA on a chromosome and inside a cell nucleus. Let’s find out what the process is and how it occurs.”
Do you know?
- DNA of a single cell length up to 2 meters.
- The distance between two base pairs is 0.34nm.
DNA (Deoxyribose nucleic acid) is a crucial, important and mysterious entity present in living organisms. Things become more complex as we explore more. Back in 1953, the chemical structure of DNA was postulated by Watson and Crick.
Since then, researchers are continuously exploring various areas of genetic research till date. A classical fact about DNA is saying, it is a ‘nucleic acid’, denoting that it is present in a cell nucleus.
Although we can’t deny the fact that DNA does exist outside the cell nucleus as well. Nevertheless, coming to the point. DNA is too tiny to see under a microscope but unfortunately, is too big to fit in a cell nucleus.
So DNA needs an organized process to fit inside a cell nucleus, organize on chromosomes and at the same time perform replication. The process of DNA packaging and de-packaging process allows the organization of DNA and replication, respectively.
Such a process is highly regulated by the interaction between DNA and proteins. The structural stability of DNA-protein assemblies are also important for the regulation of gene expression and transcriptional activities.
But you may wonder what exactly the packaging of DNA is, how it occurs, what are those proteins and what are the steps to package on a chromosome. All these and many other questions we will answer in this present article.
In this article, I will explain the theory behind DNA packing, various stages of packing, proteins and their assembly and the importance for a cell. So this article will strengthen your genetic knowledge.
“We can reach the sun and come back 300 times if we unwrap all of our DNA” blog post published on Nature Education blog by Annunziato, A. Then how is it all arranged to fit inside a cell? Let’s find out.
Key Topics:
What is DNA packaging?
As per the hypothesis, each cell contains around 2 meters long DNA. Our human body is made up of around 50 trillion cells, now imagine the entire length of our DNA. It’s insane.
However, in reality, there are some proteins like the group of histones and scaffolds that certainly help organize our DNA. By interaction with protein DNA forms, various structures coil more, supercoils on one another and prepare a compact form.
DNA supercoiling performs 5 important functions for a cell. Read this article to learn more about it.
It is interesting to understand that every time, in every cell, it forms the same structure correctly. That’s in reality again insane. So technically, the enzyme-governed process of arranging the DNA on a chromosome is referred to as DNA packaging.
It starts with the DNA-only and finally becomes chromosomes during various stages and meanwhile performs DNA metabolisms too.
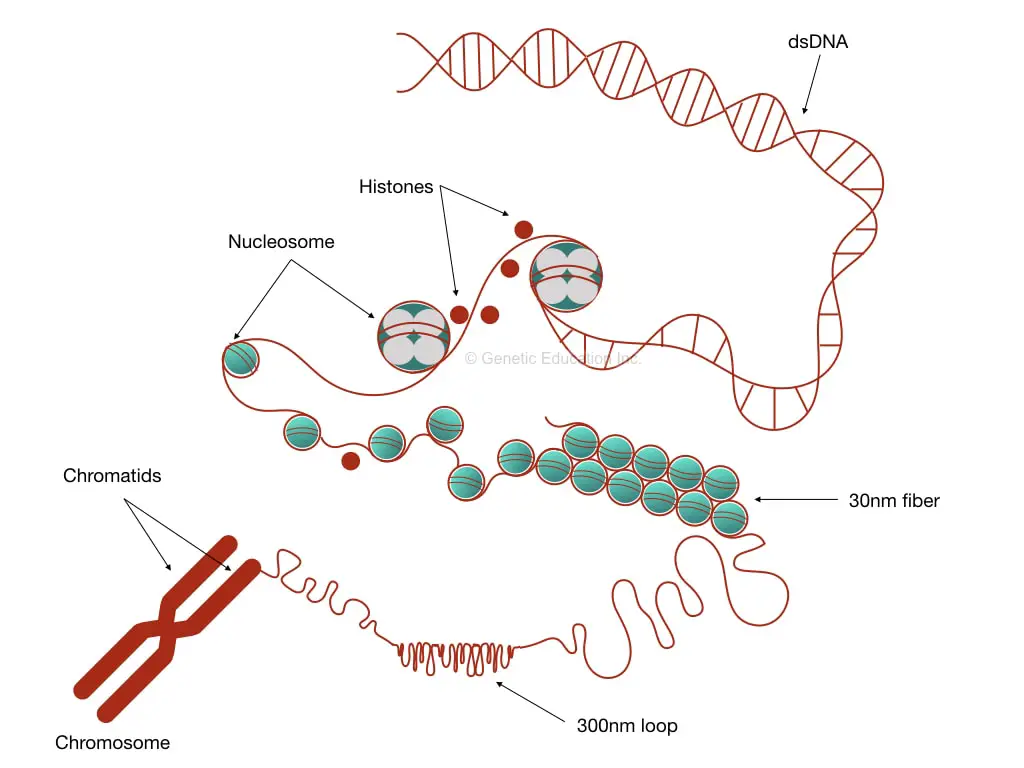
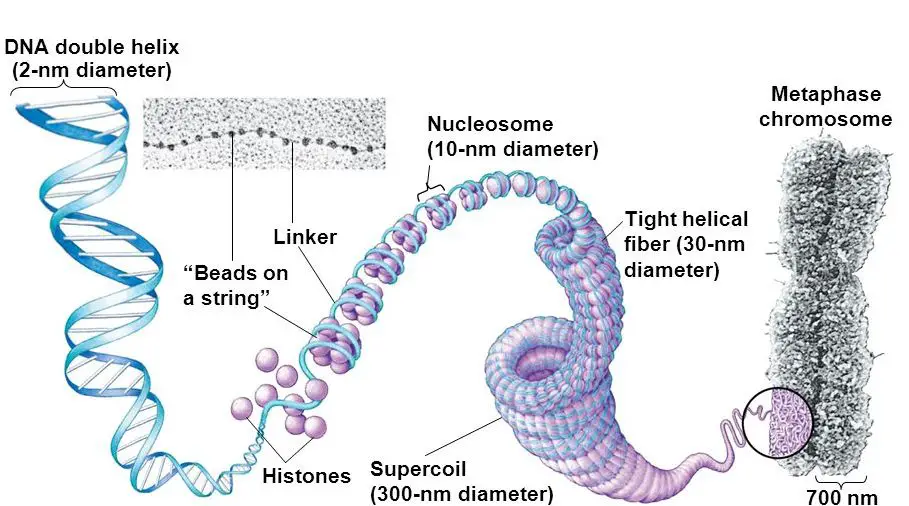
Put simply, DNA interacts with histones, forms chromatins and then a chromosome. It further coils to form 30 nm fiber and a bead-on-string-like structure. Then it becomes chromatid and finally packed on a chromosome.
Now let us understand each step precisely.
What are the steps to pack DNA on a chromosome?
The entire process of packing though includes many steps, but the basic foundation can be divided into 5 major steps. Here they are,
- Chromatin formation
- Nucleosome assembly
- 30 nm-fiber
- Chromatid
- Chromosome
How does DNA packaging occur?
Chromatin formation:
Chromatin is a simple structure, yet essential and initiates the process of DNA coiling. A group of proteins referred to as ‘histones’ and often known as DNA binding proteins are a core element of chromatin.
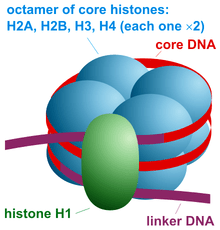
Histones are positively charged, electrostatically interact with the negative phosphate backbone of DNA and form a structure called chromatin. H1, H2A, H2B, H3 and H4 are five major histones that participate in the actual packaging process.
I have written an entire blog post on chromatin, to know more you can go and read it. Inside Chromatin: Definition, Structure, and Function.
Nucleosome assembly:
In 1974, Kornberg R and Noll M explained the nucleosome assembly in two separate studies. Kornberg R proposed the X-ray diffraction model while Noll M interpreted the visual structure.
Nucleosome assembly is a basic structural and functional unit of chromatin. It is made up of octameric protein and 146bp of DNA. The dimers of H2A, H2B, H3 and H4 form the octamer on which the DNA wraps tightly.
Note that H1 is not the part of actual core nucleosome assembly but later on participates in the process by forming a specialized structure chromatosome. Another 20 bp DNA wraps the chromatosome including the H1 and octamer assembly.
H1 histone, which is a bit larger than other histones, performs an important function to protect DNA from nucleophilic attack by binding at two separate locations. One end near the linker DNA and another between the 146bp assembly.
The additional 20 bp DNA is packed as a linker DNA which joins nucleosome assembly with each other. So overall around the nucleosome assembly and related structures, ~180-200bp DNA will be packed.
Studies demonstrate that the length of linker DNA varies from organism to organism and thus the total DNA wraps in the structure vary as well. The average length of linker DNA is 20 to 60bp.
The octameric histones are known as core assembly, core elements or core particles.
Now this entire structure Nucleosome core assembly (146bp DNA + histone octamers) + linker DNA + H1 histone is known as chromatin.
We know it’s not enough but it’s just an initiation of the process. Now thousands to millions of nucleosome assembly forms are included in a single chromosome. Moving ahead. The assembly starts forming fibers.
30 nm fiber:
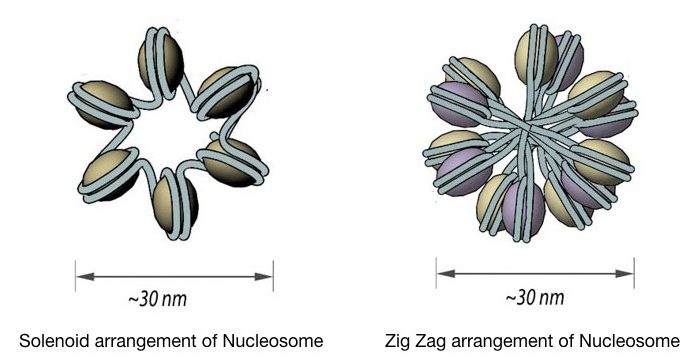
The chromatin is further coiled into even shorter, tighter, compact and thicker fiber whose length is 30 nm and so known as 30 nm fiber. It packs almost all the rest of the DNA in typically two forms, Solenoid 30 nm fiber and Zigzag 30 nm fiber.
Both models form exactly a 30nm fiber but the solenoid form is more regular in structure whereas the ZigZag form is most irregular. The spiral and regular form of solenoid includes 6 units of nucleosomes per turn face to face.
Interestingly, the Zigzag model is formed by the passing of linker DNA from the central axis which certainly does not occur in the solenoid. Some may wonder how a cell decides to become a solenoid or Zigzag fiber.
The 30nm fiber structure highly relies on the presence and the length of the linker DNA. if the linker DNA, having a variable length, as aforementioned, is long enough to pass through the central axis it becomes Zigzag or otherwise solenoid.
Here, it is very crucial to understand that the 30ng fiber is very compact and must be stabilized to remain intact. The amino acid tail of histone will remain attached to another histone unit and help stabilize the fiber.
Until now the DNA is compacted 40 folds but yet this much packaging isn’t enough to settle in a cell nucleus. Another rarely important protein often known as scaffold protein came into the picture. It loops the 30nm fiber further into nuclear scaffolds.
Chromatids:
Chromatids are the visible compact structure of a chromosome. The 30 nm fiber further coils and produces 300nm lengthed loops. Further investigations demonstrate that loops package even tighter into 250 nm wide and 700 nm lengthed chromatids.
Chromosome:
In the final stage, the chromatids form a 1400nm wider chromosome structure by joining with the centromere. The centromere of a chromosome is heterochromatin, non-coding and dark GC-rich region.
There are 23 pairs and a total of 46 chromosomes are present in a cell. Each chromosome comprises a different set of genes, coding and non-coding regions. Now when a cell enters another cell cycle, the DNA supercoiling is removed with the help of DNA topoisomerase 1 and 2 which allows DNA replication and transcription.
The tightly packed regions on a chromosome are known as heterochromatin while the loosely packed regions are euchromatin. To know more about it. Read this article: Euchromatin vs heterochromatin.
Why do we need DNA packaging?
Packaging has two crucial applications: first, as we know, it helps DNA to fit in a cell and second, it regulates gene expression. Packaging forms the most compact form of DNA and arranges it on a chromosome as we explain.
Post-translational modifications like methylation or acetylation of histones pack and unpack DNA. This means it’s a kind of switch to on or off gene expression. Tightly wrapped DNA isn’t accessible for replication and transcription.
The reason is that it doesn’t allow enzymes to settle on a substrate and perform any catalytic reaction. This means genes can’t express. On the other hand, during unwrapping, DNA becomes loosened and accessible to enzymes. Now, it can replicate and transcribe into mRNA and form a protein.
Wrapping up:
DNA packaging in eukaryotes is a complex process as compared to the prokaryotes. Packaging is as important as replication or DNA proofreading which occurs in 4 different ways.
DNA packaging is also pivotal for the regulation of gene expression. It allows and disallows chromatin to assemble. We will talk about it in some other article. There are many techniques available to understand and study DNA packaging.
Several common techniques are chromatin immunoprecipitation assay, ChIP-sequencing, DNA-protein interaction studies and methylation sequencing. Such techniques help to study how the interaction between DNA and proteins is crucial for a cell and life.
I hope this basic article with a theoretical explanation will make sense for you and helps you in your genetic learning.
FAQs:
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.



