“CRISPR naturally has spacer sequences in-between and works in sync with the CAS protein as an adaptive immune system in bacterial and archaea.”
The story of CRISPR and its fascinating applications became the ‘talk of the town’ after Doudna J & Charpentier E received the Nobel Prize in Dec 2020. Now the chatter is on its hype and obviously the stock prices of gene editing corporations too.
The scientific fraternity knows how important the present technology is and what it can do for us, but do you know it was evolved as a defense mechanism? Let me explain how CRISPR gets its life on earth and what it means for prokaryotes.
Related article: The Curious Case of CRISPR-Babies.
We get ill by acquiring viral or bacterial infections, although we have a much advanced and well-developed defense system to invade. Importantly, the same is true for all the living organisms present on earth.
Meaning, a plant or a sea cucumber may get sick when they acquire any infection. Unfortunately, bacteria face a similar scenario too. Other viruses, commonly known as “phages” infect bacteria.
They get sick too and therefore need a well-established immune system to protect themself. It took thousands of years and many evolutionary cycles for bacteria (and other prokaryotes) to develop a well-equipped, advanced, heritable defense mechanism against phages.
The newly evolved adaptive immune system isn’t so complicated but is kind of “simple but effective”; and efficient too. It has two crucial elements: a protein that destroys foreign DNA and second a storing device or memory unit that takes up some part of the foreign nucleic acid.
So basically, in the future or in upcoming generations the host bacteria can protect themself from the same infection- that’s our CRISPR CAS system. The naturally occurring bacterial defense system.
Ishino Y, Shinagawa H & Maniko K (1987) discovered the CRISPR loci in Escherichia coli for the very first time in history. During their study on iap E.coli gene, they reported an unusual repetitive and non-repetitive DNA sequence structure.
Their consideration was “nonsignificant DNA” at that time but the CRISPR element came to light after 20 years when Barrangou R & Fremaux C et al., (2007) explained CRISPR as an important element of bacterial adaptive immunity.
An unusual structure was found in the 3’- end flanking region of the iap gene. Five highly-homologous sequences of 29 nucleotides were arranged as direct repeats with 32 nucleotides as spacing. It contains dyad symmetry named REP.
Ishino Y, Shinagawa H & Maniko K (1987)
Related article: How does the CRISPR CAS9 System Distinguish Self vs Nonself DNA?- 7 Proven mechanisms.
Key Topics:
The foundation of CRISPR-CAS in bacteria:
After Barrangou R & Fremaux C et al., (2007) explained the CRISPR, ever since, the foundation of how the sequences arranged in the genome of prokaryotes get more clarity. The CRISPR sequence and genes for various CAS have been arranged in close proximity.
Spacers are located in the middle of a single CRISPR- one, two or many; depending upon the history of the host organism’s exposure to phages. That’s the key a CAS protein uses to identify the ailment agent. The CRISPR is more than just spacers, take a look at the structure.
The CRISPR loci:
Amazing information:
84% archaea and 45% bacteria consist of CRISPR as their defense system.
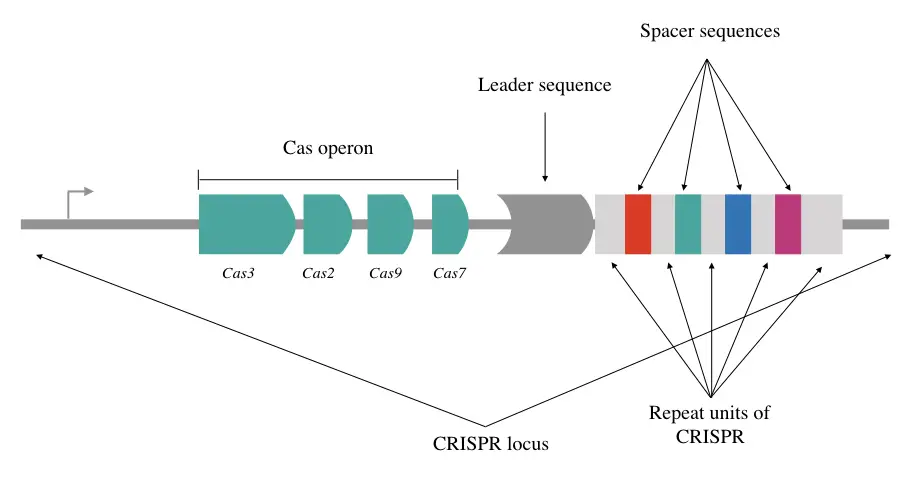
CRISPR is Clustered Regularly Interspaced Short Palindromic Repeats carrying a short repetitive sequence of 28 to 37bp. It consists of palindromically located spacer sequences which are the short and identifiable polynucleotide chains extracted from the phage or other viruses.
When the bacteria face any catastrophic viral attack it extracts some recognizable sequence and saves it in the CRISPR locus. We commonly know it as a ‘spacer sequence.’ Palindromic explains that the sequence from one end and opposite end are identical.
So the spacer sequence has significant importance for the bacterial immune system which works as a marker for future use. Studies suggest that a single CRISPR locus can have up to 100 spacer sequences, located one after another and spaced by repetitive units.
The whole system has to work in sync henceforth not only the loci carrying spacers but also the upstream and downstream region of the sequence has crucial information and importance.
For example, the leader sequences upstream to CRISPR loci and the gene for CAS in close proximity. Approx 500bp AT-rich leader sequence contains a promoter for transcription to occur and CRISPR system adaptive signaling machinery.
“The CRISPR has transmissible genetic properties and contains sequences derived from bacteriophages or plasmids.”
The CAS proteins and genes:
CAS is important for the system, as it carries the power to destroy the ds/ssDNA/RNA. We have substantial information about CAS, right!
Genes known as Cas or CRISPR-associated Cas genes, located in the CRISPR loci transcribed into protein using the leader sequence signals. The CAS protein is a simple endonuclease enzyme powerful enough to cleave the dsDNA or RNA at two separate locations.
That usually is invader nucleic acids identifiable by the crRNA formed from the spacer sequence. So overall the “in-sync” process of CRISPR and CAS destroys the ‘known’ foreign DNA and protects the primitive.
Three different types of CRISPR-Cas9 systems have been well explained in prokaryotes like bacteria and sea archaea. Among which Class I and Class II are well-studied.
The future of Gene therapy would be relying on the effective use of CRISPR technology so every single or minute detail should change the fate of the technology. Understanding the native process will become a pathbreaking comprehension for the future. Let’s go further to understand the mechanism:
How does the system work in bacteria?
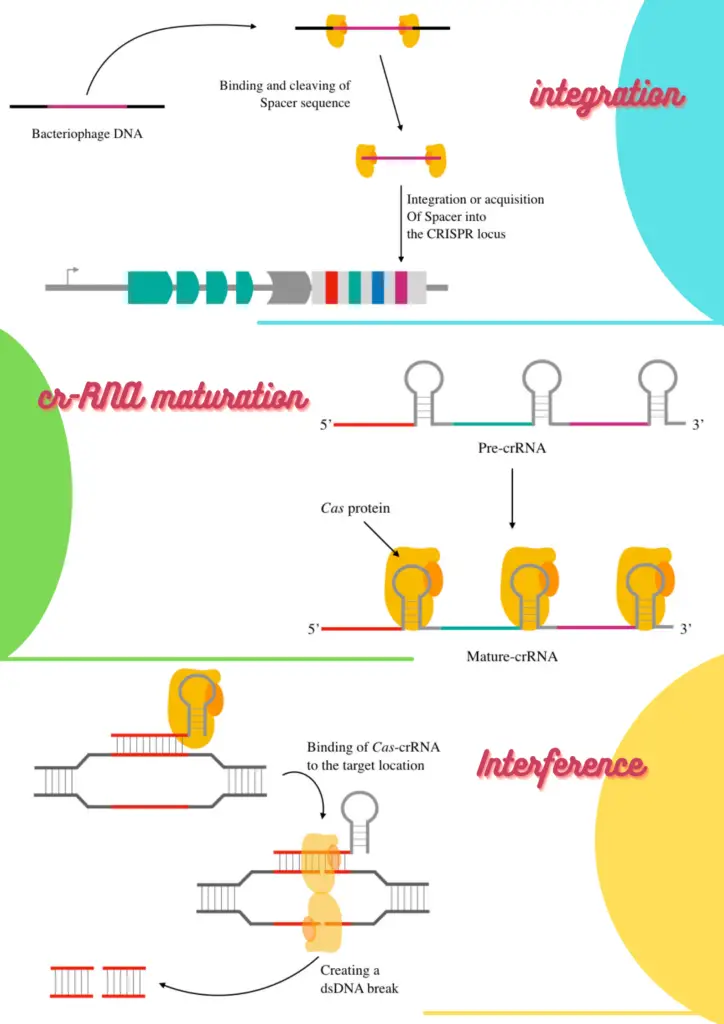
Cas genes form the protein endonuclease followed by the transcription of the CRISPR, resulting in manufacturing the “pre-crRNA”. This whole transcript is long, in fact, consists of the RNA parts of many spacer sequences.
Further in the process, it cleaved off into smaller crRNAs (depending upon each spacer sequence). Recent studies suggest that perhaps, the Cas itself performs the crRNA partition which after bindings to Cas itself directs at the target location.
When the crRNA binds its complementary location on the target or invading nucleic acid, the nuclease cleaves it off on two different locations, prevents infectivity and thereby protects the host.
Barrangou, R (2017) provided exclusive evidence, validation of the hypothesis and conferred that CRISPR provides resistance against the viral DNA. As per the literature, the process can be parted into three separate steps: Adaptation, biogenetics & processing and interference.
Do you know?
There are ten viruses per cell on the earth.
Steps in CRISPR mediated adaptive immune response:
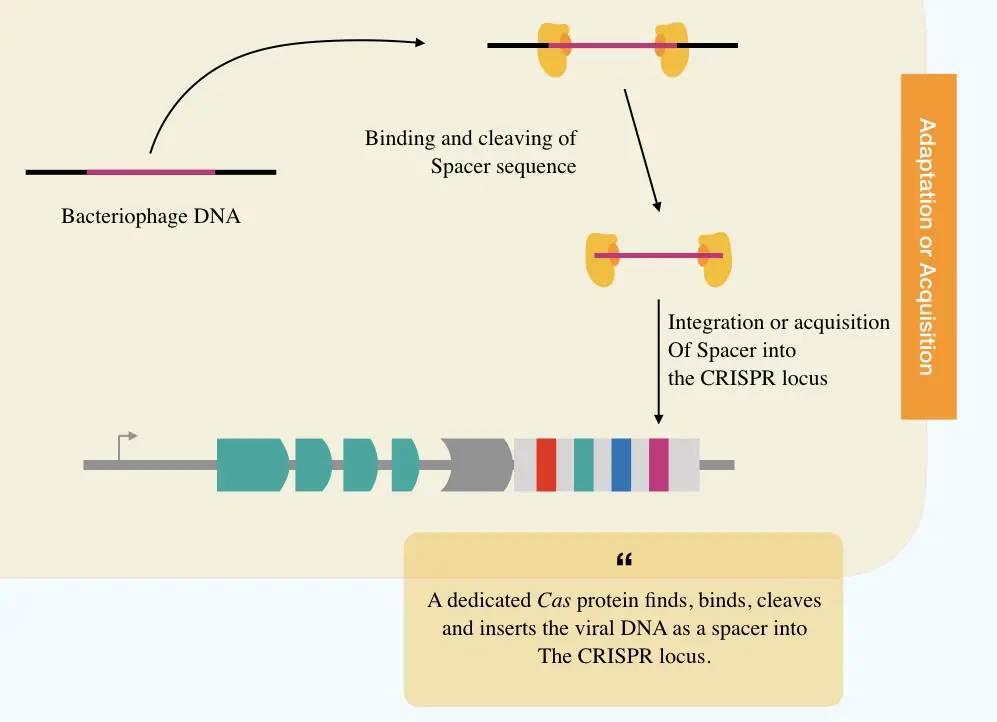
Adaptation:
The adaptation step is led by the Cas1 and Cas2 proteins which cleave off the phage DNA and initiate acquisition of the spacer into the CRISPR. Specifically, Cas1, Cas2 and Csn2 are involved in the process of spacer acquisition.
These Cas proteins are different from the ones involved in the interference process. Previous studies have provided validation that inserting mutation in any of the Cas proteins negatively impacts the activity of spacer acquisition. Some may sometimes lose function.
Note that one such mutation in the endonuclease domain inactivates the Cas for the catalytic activity known as dead Cas. Read the article here: dCAS9 (Dead CAS system): Concept, Functions and Applications.
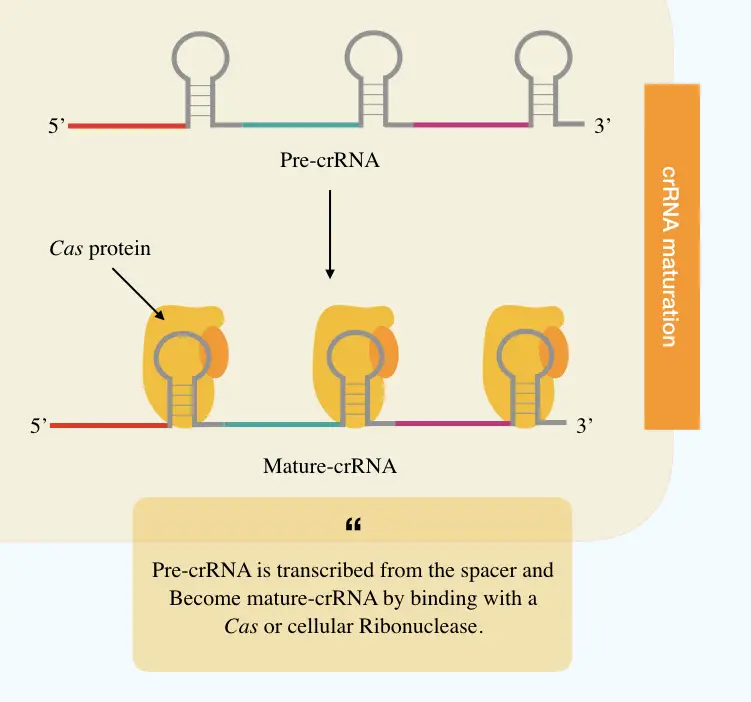
crRNA maturation:
Once the spacer gets its space into the CRISPR locus, transcription starts making the crRNA. In the enzyme governing process, first, individual spacers transcribe into the precursor crRNA or pre-crRNA, a long RNA chain.
Following it, the Cas or cellular ribonuclease protein cleaves each crRNA as per the spacer length into individual mature crRNA. Charpentier E et al., (2021) reported that a special type of tracrRNA is also present upstream of the CRISPR loci and co-processed when paired with the crRNA.
Their findings also suggest that the Cas9 protein provides the complementary binding between tracrRNA and crRNA for more specific interference. Moreover, any acquired mutation in Cas9 protein prevents duplex formation.
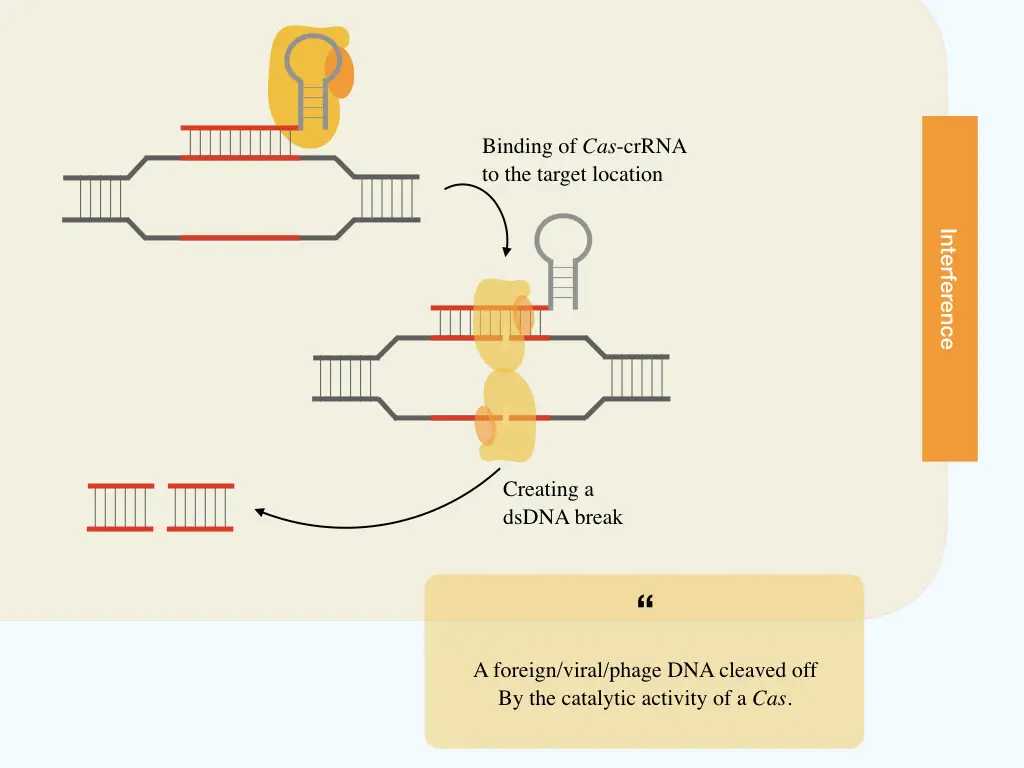
Interference:
In the last and final step, the mature crRNA complex (crRNA and Cas9) migrates toward the foreign or phage nucleic acid. Here the crRNA works as a guide for Cas9 to find the exact target location or sequence to cleave.
Once the hassle of finding the invader completes, the crRNA binds to the complementary location followed by localizing of Cas9 to two strands of the dsDNA. Once the DNA binding domain is attached to each stand, the catalytic domain breaks the phosphodiester bonds and the dsDNA, afterward.
Resultantly, the phage DNA can’t metabolize into the host and is eventually destroyed. Nonetheless, the process may vary a bit from system to system.
Note:
The tracrRNA has an important role in crRNA maturation and interference. The Nobel Prize-winning study suggests that tracrRNA-crRNA collectively accurately guides the Cas9 and is hence referred to as sgRNA or guided RNA. I will cover a whole article on this topic later.
CRISPR systems in bacteria:
The adaptive immune system almost remains the same in bacteria but may vary from archaea, however, therefore several types of CRISPR systems are reported in various prokaryotic bacteria.
Majorly those are classified into class 1 and class 2 in which types I, III, IV belong to class 1 while II, V and VI belong to class 2. Note that the classification system relies on the effector molecules of CRISPR.
For instance, the class 1 CRISPR system consists of multiple subunits of effector proteins while the class 2 CRISPR system majorly consists of a single large effector protein.
This section of the article mainly focuses on the concept of what a different CRISPR system is and why it’s important.
Class I CRISPR system:
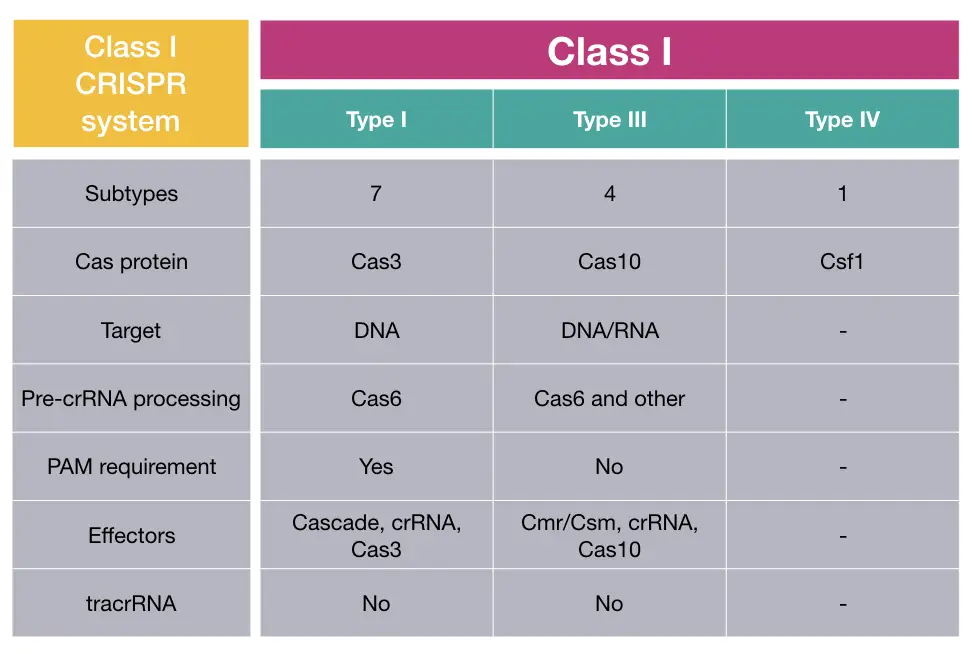
The class I system is prominently found and is the most common and present in 90% population of bacteria and archaea. It is further divided into types I, III, IV and sometimes 12.
The effector complex of the present system is multisubunit in nature and possesses signature nuclease protein Cas3, Cas10 and Csf1 in type I, III and IV, respectively. DNA and RNA both are common targets here.
Studies suggest that the clear target for the Csf protein (type IV) has yet not been studied. Cas3 and Cas10 have DNA as their main target.
Contrary, functional studies also explain that the Cas3 and Cas10 from systems I and III perform ssDNA cleavage and nascent RNA binding functions but the function of multisubunit Casf1 remained remarkably unclear from type IV.
The class I type I has been well-established and studied and suggests that it required Cas6 protein and PAM (Protospacer Adjacent Motif) sequence for pre-crRNA processing and target recognition, respectively.
The Cas6 performs pre-crRNA processing in type III as well, but it needs other proteins as well to achieve precision. Notedly, PAM sequences aren’t necessarily needed and henceforth, hypothetically less effective in comparison to type I.
The type IV of class I CRISPR system is less studied as its target, pre-crRNA processing protein and recognition domains are yet not reported. TracrRNA isn’t required for the class I CRISPR system.
Class II CRISPR system:
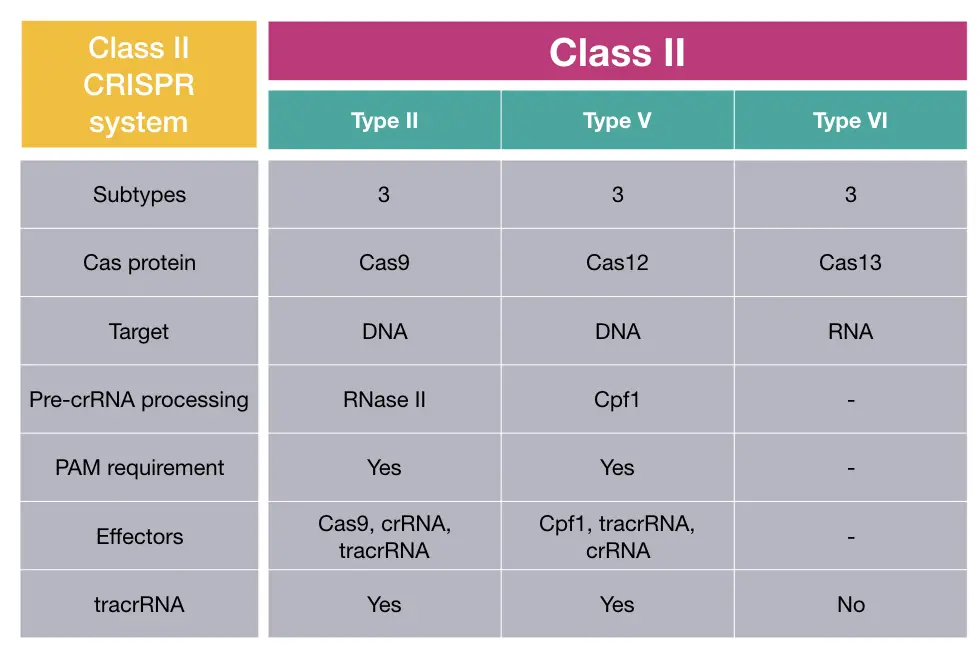
The class II CRISPR system is categorized based on the presence of a single, large prominent, effector protein, and has further been classified into 3 types: Type II, Type V and Type VI.
Both DNA and RNA are common targets for endonucleases of the class II CRISPR system. Importantly it’s only present in bacteria.
Cas9, Cas12 and Cas13 are common endonucleases found in type II, V and VI, respectively. Each one has 3 subtypes as well, among which type VI only needs an RNA target. DNA is the common target for Cas9 and Cas12 of Class II and V.
The protein required for pre-crRNA processing in classes II and V is RNase III and Cas12. The crRNA processing mechanism and involved protein remained nuclease for class VI.
Further to this, type II and V rely on the presence of PAM sequence for recognition and cleavage which is also not required for class VI.
Research on RNA processing indicated that the tracrRNA is required for type II and V, unlike class I.
| Type I | Type III | Type IV | Type II | Type V | Type VI | |
| Subtypes | 7 | 4 | 1 | 3 | 3 | 3 |
| Cas protein | Cas3 | Cas10 | Csf | Cas9 | Cas12 | Cas13 |
| Pre-crRNA processing | Cas6 | Cas6 and other | – | RNase III | cpf1 | – |
| Target | DNA | DNA/RNA | – | DNA | DNA | RNA |
| Requirement of PAM | Yes | No | – | Yes | Yes | – |
| Requirement of tracrRNA | No | No | – | Yes | Yes | – |
So what’s a catch?
CRISPR gained special attention after 2020 (obviously because of the Nobel Prize-winning findings!) But what’s the catch? Why has it gained so much attention? The reason is simple, It is an effective gene-editing tool.
As we talked, the mechanism works around the CRISPR locus, Cas protein and crRNA. Suppose if we replace the spacer sequence with the gene of interest or sequence or exon we wish to incorporate into the genome. What will happen?
The Cas identifies it as a spacer and tries to incorporate it as correctly as it can, and it’s done. We want to do exactly that. And the rest- how it works we all know.
It has tremendous applications in gene editing, gene expression, monitoring cell activity, improving yield and achieving the desired phenotype.
This article will help you to learn more: Applications of CRISPR-CAS9 in Medical Science, Diagnostics, Research, Plant Biology and Agriculture.
A study conducted by Synthego explained that 45% of commercial and 48% of non-commercial organizations use CRISPR technology. Note that RNA interference technology is on the second number of the chart. Take a look at the chart here:
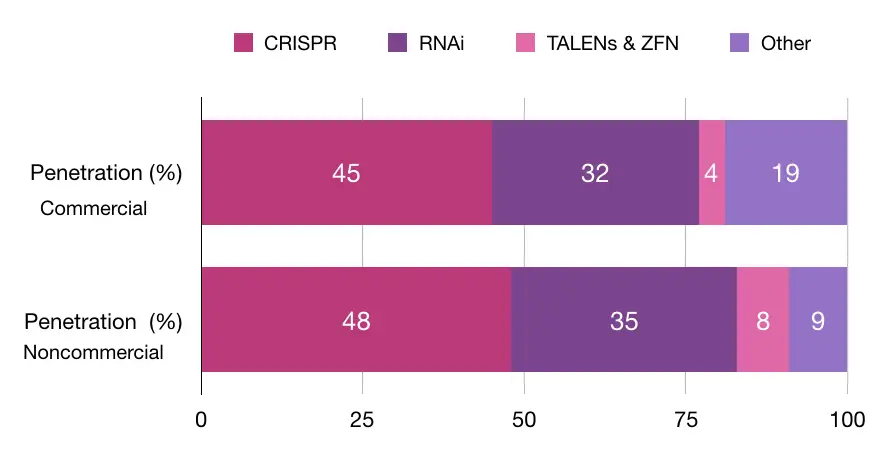
In the same study, other data suggest that gene knockout is one of the most popularly used using the CRISPR technique. Take a look at here:
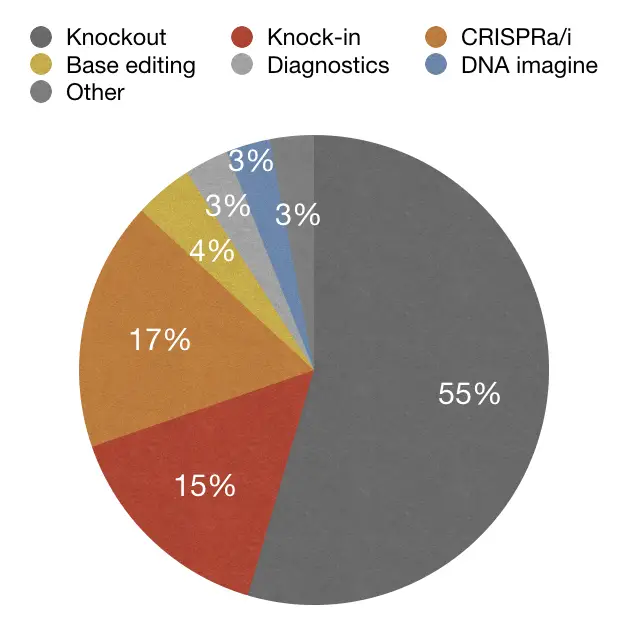
So the data-driven studies suggest that, Yes, It will be the future of gene therapy and has lucrative applications and excellent precision.
This information has been extracted from this article: State of CRISPR Gene Editing in the Drug Discovery sector. We are thankful to the publisher and compny to provide such important and valuable information.
Wrapping up:
Evolution is a “must-need” process for life on earth to survive. CRISPR is an adaptive immune system, prokaryotes developed for their own protection. Thanks to them, we would make our future bright.
We can edit genes, repair genes, cure disease, prevent genetic abnormalities, create more GMOs, improve the economy, create more diverse plant and bacterial species and much more.
However, keep in mind that the technology is still growing, not among the common methods used in most labs and that’s why it is tedious, time-consuming and costly.
This article is my small attempt to explain how things work naturally and I hope you people understand it. In addition, I also want to tell you that if you are a life science student, please start learning and doing research around CRISPR and gene editing because it’s our future.
I hope this article makes sense to you.
Resources:
Loureiro A, da Silva GJ. CRISPR-Cas: Converting A Bacterial Defence Mechanism into A State-of-the-Art Genetic Manipulation Tool. Antibiotics (Basel). 2019;8(1):18. Published 2019 Feb 28. doi:10.3390/antibiotics8010018.
Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37(1):7-19. doi:10.1016/j.molcel.2009.12.033.
Newsom, Sydney. (2021). The CRISPR-Cas Mechanism for Adaptive Immunity and Alternate Bacterial Functions Fuels Diverse Biotechnologies. Frontiers. https://www.frontiersin.org/articles/10.3389/fcimb.2020.619763/full.
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429-5433. doi:10.1128/jb.169.12.5429-5433.1987.
Barrangou, Rodolphe, et al. “CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes.” Science, 23 Mar. 2007, https://www.science.org/doi/10.1126/science.1138140.
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014 Nov 28;346(6213):1258096. doi: 10.1126/science.1258096. PMID: 25430774.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.