“Learn the basics of short tandem repeats (STRs), why we use them in DNA profiling and their applications in this article.”
Our genome is mysterious! Where only a small portion of the genome can encode proteins, a large portion (97%) is non-coding. Repetitive sequences, transposons, introns and non-coding genes are commonly found as non-coding DNA.
Among all these, the largest component of our genome is (non-coding) repetitive DNA sequences. VNTRs and STRs, popularly known as mini- and microsatellites, respectively are repetitive elements of our genome.
Variable Number of Tandem Repeats and Short Tandem Repeats both are types of tandem repeats, located one after another in a sequence or at a particular genomic location. Interestingly, both VNTRs and STRs are utilized due to their polymorphic characteristics.
You’ve likely come across STRs in various discussions or studies, given their widespread use in DNA typing. But do you know more about STR? How exactly it’s used in DNA typing?
In this article, I will explain to you the concept of STRs, what they are, their nomenclature, why we use them and their applications.
Stay tuned.
Related article: DNA Fingerprinting- Definition, Steps, Methods and Applications.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension.
Key Topics:
What are STRs?
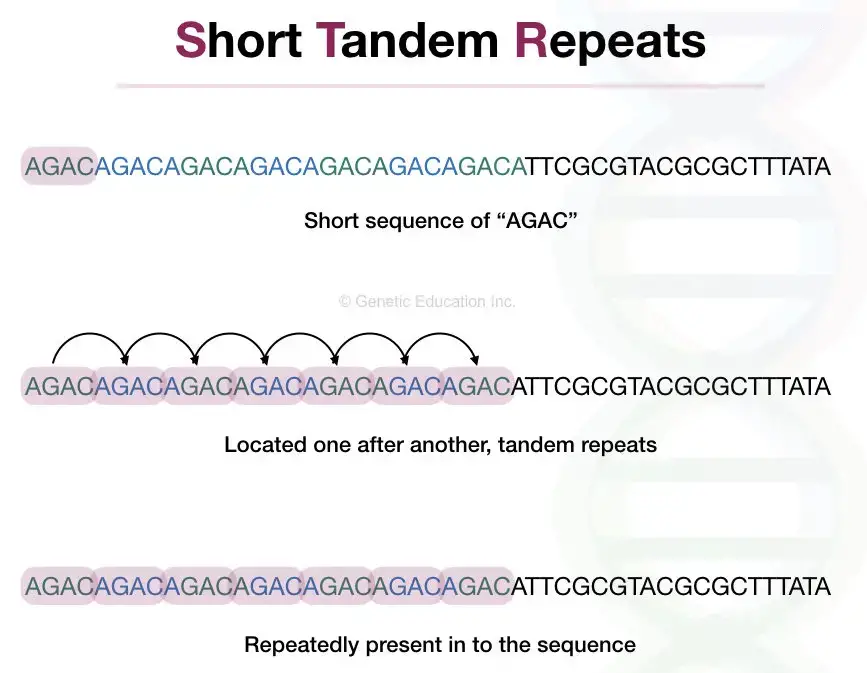
STR stands for Short Tandem Repeats. STRs are short repetitive DNA units of 1 to 6 nucleotides, present up to 50 times in a sequence and lengthed 100 to 120 bp. They are non-coding DNA that can not participate in protein synthesis.
The present short repeats are mostly scattered on the telomeric regions of chromosomes and, hence, are also known as microsatellites. It comprises 3% of the total genome and is present in prokaryotes and eukaryotes including humans.
Notably, among the 3 to 4% of the total STR portion, a small number of STRs (8 to 10%) are located in the coding regions and genes. Studies suggest that STRs are highly rich in “Adenines.”
Short = ranging from 1 to 6 nucleotides., Tandem = located one after another at a specific location and Repeats = repeatedly present at a specific interval.
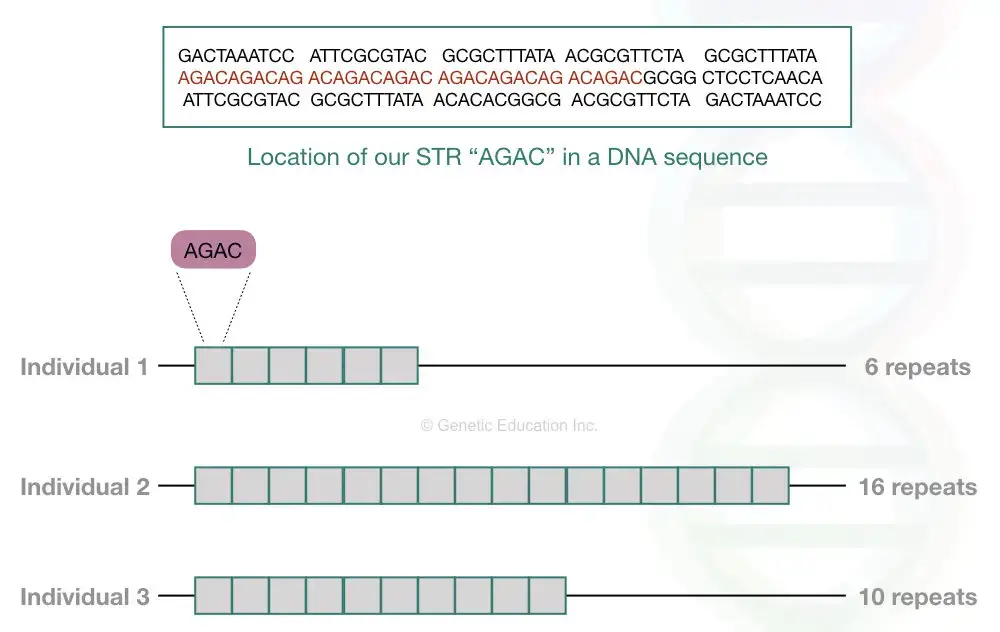
Each STR varies in the number of repeats and nucleotides it contains. For instance, if an STR consists of two nucleotides, it’s termed dinucleotide repeats; for three nucleotides, trinucleotide repeats, and so forth. Refer to the table below for more details.
| Repeat Type | Examples |
| Dinucleotide (2) | (CA)n, (GT)n, (AG)n, (TG)n |
| Trinucleotide (3) | (CAG)n, (AGC)n, (GAA)n, (TGC)n |
| Tetranucleotide (4) | (AAAT)n, (AGAT)n, (ATGC)n, (TGCA)n |
| Pentanucleotide (5) | (AAAGC)n, (TATAT)n, (CGCGC)n, (GAGCT)n |
| Hexanucleotide (6) | (AGATCT)n, (TCTAGC)n, (GAAACC)n, (CACAGG)n |
Notedly, tetra-, penta- and hexanucleotide repeats are extensively used in various DNA typing experiments, most significant STRs are dinucleotide repeats.
Interestingly, any single STR is present after every 2000 nucleotides in our genome, whereas chromosome 19 comprises the highest number of STR markers. Based on the repeat structure, STRs are further classified into simple sequencing repeats and compound repeats.
The simple sequencing repeat contains only a single type of repetitive unit in the STR. Examples of simple repeats are D5S818, D13S317, D16S539 and TPOX.
>> For instance, CACACACACACA.
Contrarily, compound STR repeats contain a combination of various repeats. An example of a complex or compound STR is D21S11.
>> For instance, CA CA AGATCT CA AGATCT AAAT AAAT CA.
Usually, because of the less sequence complexity, simple sequence repeats, particularly the penta and hexanucleotide repeats have been used in DNA typing experiments.
STR Nomenclature
If you are a bit familiar with STRs you know that they are denoted as D13S317, D17S30 or D1S80, etc. These are the unique IDs, given to each STR. The DNA Commission of the International Society of Forensic Haemogenetics proposed the nomenclature method of STRs (ISFH) in 1993.
For D13S317, the D is for DNA which is located on chromosome number 13. S is for the STR sequence and 317 (in D13S317) and 30 (in D17S30) are the unique sequence numbers.
Although the present nomenclature method is not uniform, other markers are named differently. For example, APOB, APOA2 and CYAR04, etc.
Examples of STRs:
The present list of STRs represents various core and other STR markers used in forensic and paternity testing or typing. The table shows the example of markers, their chromosomal location, STR repeat unit and mutation rate in percentage.
| Marker | Chromosomal Location | STR Repeat Unit | Mutation rate (%) |
| D3S1358 | 3p21.31 | (TCTA)n | 0.12 |
| TH01 | 11p15.5 | (AATG)n | 0.01 |
| D5S818 | 5q23.2 | (ATCT)n | 0.11 |
| D7S820 | 7q21.11 | (TATC)n | 0.1 |
| D8S1179 | 8q24.13 | (TCTA)n (TCTG)n | 0.14 |
| D13S317 | 13q31.1 | (TATC)n | 0.14 |
| D16S539 | 16q24.1 | (GATA)n | 0.11 |
| CSF1PO | 5q32 | (ATCT)n | 0.16 |
| Penta E | 15q26.2 | (AAAGA)n | 0.16 |
| D18S51 | 18q21.3 | (AGAA)n | 0.22 |
| vWA | 12p13.31 | (TAGA)n (CAGA)n | 0.17 |
| D21S11 | 21q21.1 | (TCTA)n | 0.19 |
| D2S1338 | 2q35 | (GGAA)n (GGAC)n (GGCA)n | 0.12 |
| D19S433 | 19q12 | (CCTT)n ccta (CCTT)n cttt (CCTT)n | 0.11 |
| TPOX | 2p25.3 | (AATG)n | 0.01 |
| D12S391 | 12p13.2 | (AGAT)n (AGAC)n | 0.17 |
| D1S1656 | 1q42.2 | (CCTA)n (TCTA)n | <0.25% |
| D6S1043 | 6q15 | (AGAT)n | – |
| D22S1045 | 22q12.3 | (ATT)n act (ATT)n | <0.25 |
| FGA | 4qq31.3 | (GGAA)n (GGAG)n (AAAG)n (AGAA)n (AAAA)n (GAAA)n | 0.28 |
| D10S1248 | 10q26.3 | (GGAA)n | <0.25 |
| D2S441 | 2p14 | (TCTA)n | <0.25 |
Why are STRs Important?
Now, let’s see why STRs are important and used for various DNA typing experiments. First, what is DNA typing?
DNA typing is a technique to study the DNA for identification purposes. For instance, paternity DNA testing, maternity DNA testing, forensic analysis, and biological identifications.
Here are a few reasons why STRs are important for DNA typing and Kinship testing.
Abundance: STRs are ubiquitous in every organism on earth from prokaryotes to eukaryotes. Humans also contain various STRs on different chromosomes with different repeat numbers.
Codominance inheritance: The codominance inheritance characteristic of STRs allows a unique combination of new alleles in a population. Thus it creates new allelic diversity and helps in investigating biological identity.
Heterozygosity: each separate allele is inherited from an individual parent. This will provide the power to distinguish heterozygosity alleles. Meaning, a distinct heterozygous pattern helps in accurate investigation.
Practically, we observe two separate alleles as DNA bands in a gel or capillary electrophoresis.
Highly polymorphic: STRs are chosen for various DNA typing procedures because of their highly polymorphic nature. Therefore, there is a one-in-a-billion chance that two individuals have the same STR profile for a given set of STRs (16 STR markers as per the standard).
High mutation rates: STRs demonstrate remarkably high mutation rates, leading to the generation of new alleles and variations within the population. This phenomenon produces allelic polymorphism and novel genetic diversity.
Notedly, the typical mutation rate per locus per generation is 10 ^(-3) to 10 ^(-6).
Location: STRs are typically present in the non-coding part of the genome. Previously it was believed that the present genomic portion is non-function, however, it regulates gene expression. STRs used in DNA typing can not influence gene function.
Efficient amplification: For PCR-based and DNA sequencing-based STR typing, amplification is pivotal. Interestingly, as the STRs are more rich in A and AT-rich sequences, it’s comparatively easy to design and perform PCR amplification.
Streamlined procedure: STR fragments typically range from 100 to ~500 bp. The short fragment size makes it feasible for PCR amplification, sequencing and fragment analysis. So STR typing experiments are streamlined and standardized.
Availability: These are the common reasons, STR typing is most prevalent, having various ready-to-use kits, components, reagents, assays and instrumental setups available in the market.
These are the common reason, STRs are widely used in DNA typing and testing experiments.
Types of STRs
Based on the location, STRs are classified into autosomal STRs, located on the autosomes and sex-chromosome STRs, located on X and Y chromosomes.
Autosomal STRs:
Autosomal STRs follow the classic Mendelian inheritance pattern and are located on chromosomes 1 to 22 (non-sex chromosomes). This means that each STR allele is inherited from both parents.
Due to the highly polymorphic nature of autosomal STRs, and identification power regardless of gender, it’s extensively used in forensic investigations, paternal verification and population genetic studies.
The codominance and highly polymorphic nature also provide unique allelic combinations, variations and repeat numbers. Common autosomal STR markers used and approved by International authorities are D3S1358, vWA, D16S539, D2S1338, etc as core STR markers.
In addition, TH01, D21S11, and FGA are other additional STR markers also included in various forensic and kinship testing. Autosomal STRs show negligible chances of genetic similarity.
Sex-chromosomal STRs:
The sex chromosomal STRs are located on sex chromosomes. I.e X and Y chromosomes. In addition to genetic relatedness investigation, it also provides information on one’s sex or gender.
Unlike the autosomal STR inheritance pattern, sex chromosomal STRs follow different inheritance patterns for males and females. Y-chromosomal STRs are inherited in male individuals only while X-chromosomal STRs can be inherited in males and females.
Particularly the Y-STRs are less polymorphic and can not be used for routine kinship and forensic investigations. It can only be used for tracing parental lineage and male-specific identification.
Contrary, the X chromosomal STRs can be used for complex kinship investigations but not for tracing the maternal lineage as it can not be exclusively inherited from the mother.
For tracing the maternal lineage, Mitochondrial DNA test as mt-DNA typing in forensic studies is used.
Commonly used Y-STR markers are DYS391, DYS385, DYS19, and DYS458 and X-STR markers are DXS10103, DXS8378, and DXS7132.
Related article: What If the Y chromosome Completely Vanished From Earth?
What are STRs used for?
Now, the important part of this article is the applications of STRs. In this section, we will understand STRs used in various fields.
Forensic DNA profiling:
STR is extensively used by various forensic organizations for criminal verification and identification. The investigator matches the STR profiling data of the crime scene with the reference data or suspect’s DNA data.
Commonly 20 to 25 markers have been used for forensic STR profiling. As per the NIST (National Institute of Standards and Technology) database, 20 markers are considered as CODIS (Combined DNA Index System) core STRs.
Refer to this table to know the number of STRs used for different investigations.
| Application | Number of STRs | List of STRs |
| Forensic Analysis | Typically 20-25 markers | D3S1358, TH01, D5S818, D7S820, D8S1179, D13S317, D16S539, CSF1PO, Penta E, D18S51, vWA, D21S11, D2S1338, D19S433, TPOX, D12S391, D14S306, D1S1656, D6S1043, D22S1045, FGA, D10S1248, D9S1122, D2S441, D17S1301 |
| Paternity Verification | Typically 15-20 markers | D3S1358, TH01, D5S818, D7S820, D8S1179, D13S317, D16S539, CSF1PO, D18S51, vWA, D21S11, D2S1338, D19S433, TPOX, D12S391, D14S306, D1S1656, D6S1043, D22S1045, FGA |
| Genetic Relationship Study | Varies depending on the study | D19S433, vWA, TPOX, D12S391, Penta E, Penta D, etc. |
| Population Genetics | Varies depending on the study | CSF1PO, THO1, D6S1043, D11S2010, D14S306, D20S482, etc. |
However, the number of the core and all the markers may vary among countries based on their standards.
Note: Along with these STR markers Mitochondrial markers are also used for forensic typic.
Paternity Testing or Typing:
Yet, another extensive application of STRs is in paternity testing, child custody and kinship testing. With utmost precision, a conclusive biological relationship can be established between putative parents (father or mother, or both) with the child.
To do so various STR markers are matched between parents and child using sophisticated techniques like capillary gel electrophoresis. STR-based paternity typing gives 99.99% accurate biological identification.
As per the international standards 16 core STR markers are used for kinship and paternity typing. However, testing facilities usually recommend testing 21 markers to increase the precision and reduce the likelihood of errors.
Ancestry DNA testing:
The widely popular ancestry DNA testing which tests 7,00,000 different markers across the genome, also tests for STR markers as well. Usually, Y-STR markers have been included in these tests to establish paternal lineage.
However, ancestry DNA testing does not entirely rely on the STRs, they also test for mitochondrial markers and various SNPs.
Genetic maps:
Thousands to millions of STRs are located across the genome on various chromosomes. Those can be used to prepare a genetic map.
Population genetic studies:
STR markers have been a significant tool for population genetic studies. Various STRs are tested from various populations to determine the allelic frequency for each STR allele.
After that, the allelic frequency variation has been used to study population diversity, structure, and genetic relationship with other human populations. Furthermore, migration patterns and historical demographical events like anthropological studies are also conducted.
Genetic disease studies:
Previously it was believed that STRs are located only on non-coding DNAs and do not have any significant role in disease development. However, research studies showed that 8 to 10% of STRs are also located in coding regions and cause various genetic conditions.
For example, various types of Fragile X are caused by CCG/CGG triplet repeat expansion present on the X chromosome. Here is a short list of various disorders caused by STR expansions.
| Disease | Gene | Chromosomal Location | Repeat Unit |
| Huntington’s disease (HD) | HTT | 4p16.3 | CAG |
| Myotonic dystrophy type 1 | DMPK | 19q13.32 | CTG |
| Fragile X syndrome | FMR1 | Xq27.3 | CGG/CCG |
| Friedreich’s ataxia | FXN | 9q13-q21 | GAA |
| Spinocerebellar ataxia 1 | ATXN1 | 6p22.3 | CAG |
Abnormal expansion of STR repeat on different genes causes various genetic disorders. Moreover, recent studies also showed that STR mutations are also directly linked with gene expression and regulations.
Comprehensive research is required to shed light on STR’s role in gene expression and regulations.
Detection of maternal cell contamination:
Maternal cell contamination is a common problem scientists face during prenatal genetic studies. Here, the fetal cells (amniotic fluid or chorionic villi) can be contaminated with maternal cells during sample collection.
Using the combination of STR and VNTR markers MCC can be detected effectively.
Other applications:
In addition to this, STR typing has been used in chimerism and genealogy studies, cell line validation and linkage analysis.
Wrapping up:
STRs are great tools for human biological identification. It will give 99.99% accurate results. Using a different combination of various STRs, the chances of genetic relatedness can be reduced by 1 in a billion.
However, the complex structure of several STRs makes it difficult to use them. Still, the combined 20 to 30 markers are sufficient enough for various DNA typing studies.
I hope you like this article. In the next article, we will discuss various types of STR typing techniques and analysis. Share this article and subscribe to our blog.
Sources:
Fan H, Chu JY. A brief review of short tandem repeat mutation. Genomics Proteomics Bioinformatics. 2007 Feb;5(1):7-14. doi: 10.1016/S1672-0229(07)60009-6. PMID: 17572359; PMCID: PMC5054066.
Database of short tandem repeats by stripy.org.
FBI CODIS core STR loci by NIST.STR database by NIST.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.



