“Replication is an enzyme governed process, in which new DNA molecule is formed by the process of semi-conservative DNA replication.”
DNA is everything to us, meaning it’s a building block of life. Everything starts and ends at DNA. So it is important for us. Functionally active DNA sequences are genes having a role in making proteins.
The DNA is a double-helix structure with a phosphate backbone, sugar and nitrogenous bases, and is known as deoxyribonucleic acid. It’s a kind of nucleic acid present in the cell nucleus.
DNA functions to tailor protein and transmit from parent to their offspring. Therefore “inheritance” is one of its important characteristics.
To function correctly every time, new DNA should be formed, every time, during every cell cycle. The biological process of making a new DNA or DNA strand is referred to as “replication”.
DNA synthesis– making a new DNA, is an enzyme governed catalytic reaction involving DNA polymerase, ligase, helicase, topoisomerase, etc. DNA polymerase has a significant role in synthesizing new DNA by incorporating dNTPs into the growing strand.
I was confused while writing this article, the reason is the topic “replication” itself. We can explain it in a single paragraph or a whole book isn’t enough to explain. In this article, I have tried to explain it and hope it will be understandable to all groups of students.
This article is a brief overview of how replication occurs in a cell. It’s a kind of educational article, we can say, but is written in simple language.
Common terms used:
| Term | Explanation |
| Replication | The process of synthesis of DNA. |
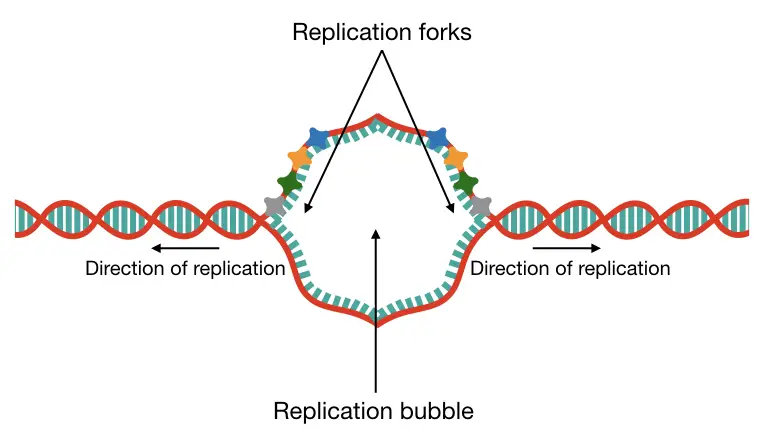
| Replication fork | The Y-shaped structure is formed when helicase unwinds the dsDNA. |
| Origin of replication | From where does the polymerase start replication |
| Replication bubble | When helicase unwinds a single dsDNA strand in opposite direction. |
| Chromosome | A complex network of DNA and protein. |
| Helicase | Unwinds the DNA, convert dsDNA into two ssDNA |
| Topoisomerase | Releases the tension on DNA. |
| Polymerase | The enzyme synthesizes a new strand of DNA. |
| Primase | Synthesize RNA primer |
| Ligase | Fills gaps between adjacent nucleotides |
| SSB protein | The single-stranded binding protein binds to the single-strand and prevents the rebinding of DNA. |
| Leading strand | The ssDNA from where the DNA replication start |
| Lagging strand | The ssDNA from where the DNA replication ends. Replication occurs in fragments. |
| Okazaki fragments | Short DNA fragments facilitate replication on lagging strands. |
Key Topics:
What is DNA replication?
Replication is a process of copying the DNA; exactly similar dsDNA molecule forms. Prokaryotes and eukaryotes have a similar mechanism of nucleic acid synthesis, involving polymerase as a key enzyme.
Put simply, replication occurs during cell division. The DNA polymerase incorporates nucleotides when dsDNA unwinds and becomes single-stranded.
Read more: Prokaryotic DNA Replication: Initiation, Elongation and Termination.
Definition:
The in vivo process of copying or synthesizing the DNA using the DNA polymerase is known as replication.
The semi-conservative mode:
Eukaryotes and higher in the newly formed duplex prokaryotic organisms follow a specialized model or synthesis known as the semi-conservative in the newly formed duplex mode of replication. The model best explains the replication process precisely.
Meselson M and Stahl F have explained the semiconservative nature of replication in 1985. The model is explained here;
DNA is double-stranded, we all know, in this mode of replication, when two strands separate, each work as a template for replication to occur. Although replication progression is different for each strand.
Each serves as a template substrate for DNA polymerase to synthesize a new strand, meaning in the end, four different single-stranded DNA forms- two dsDNA One
One is our parental DNA strand while the newly synthesized is our daughter strand. All 4 ssDNA replicates to make 8 ssDNA and so on. We will explain the whole process of semi-conservative mode in some other articles. Coming back to the point- replication.
The process of DNA replication:
The process of DNA replication is best explained using the E Coli model organism for both prokaryotes and eukaryotes. It includes three steps: Initiation, elongation and termination. Before initiation, a replication fork forms to start the process.
Several proteins, enzymes and a short RNA are involved in the process helping to form DNA. Replication is important for reproduction, cell growth and DNA repair.
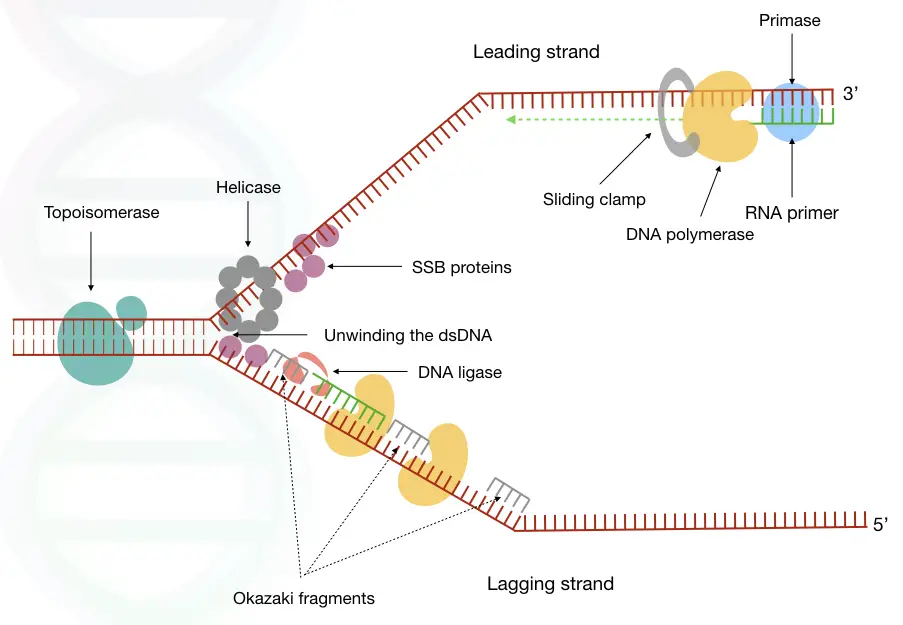
Steps in DNA replication:
Initiation, elongation and termination are three steps in DNA replication. Detail of every step explained here:
Initiation:
The process starts when the DNA is ready to be a part of the part. DNA isn’t normally available for replication, because of DNA packaging. Histones interact with DNA and wrap it in nucleosomes, chromatin and chromosomes.
Chromosomes have tightly wrapped, compact DNA threads, and aren’t accessible to replicate. Certain proteins unwind DNA and make them available. A pair of helicases is first recruited to the site of replication.
Helicase’s function is to unwind the dsDNA into two single-stranded ones. It forms a Y-shaped structure, known as “replication fork”, from where the replication starts.
The junction of the Y-shaped structure that allows the polymerase to replicate is referred to as the “origin of replication”. It’s a specific location in the eukaryotic genome having a specific kind of DNA sequences to recognize by the enzyme, for example, consensus sequences.
The origins of replication are mostly A/T rich sequences.
More than 1,00,000 origins of replication occur at once on a single human chromosome.
Unfortunately, those aren’t present in our genome. Consensus sequences are commonly present in yeast. Replication is a bidirectional process, meaning, two different helicases work in opposite directions to unwind the DNA.
Consequently, two different replication forks and origins of replication form and create a replication bubble (see the image). Many different replication bubbles are formed to initiate replication at thousands of locations on a chromosome.
Elongation:
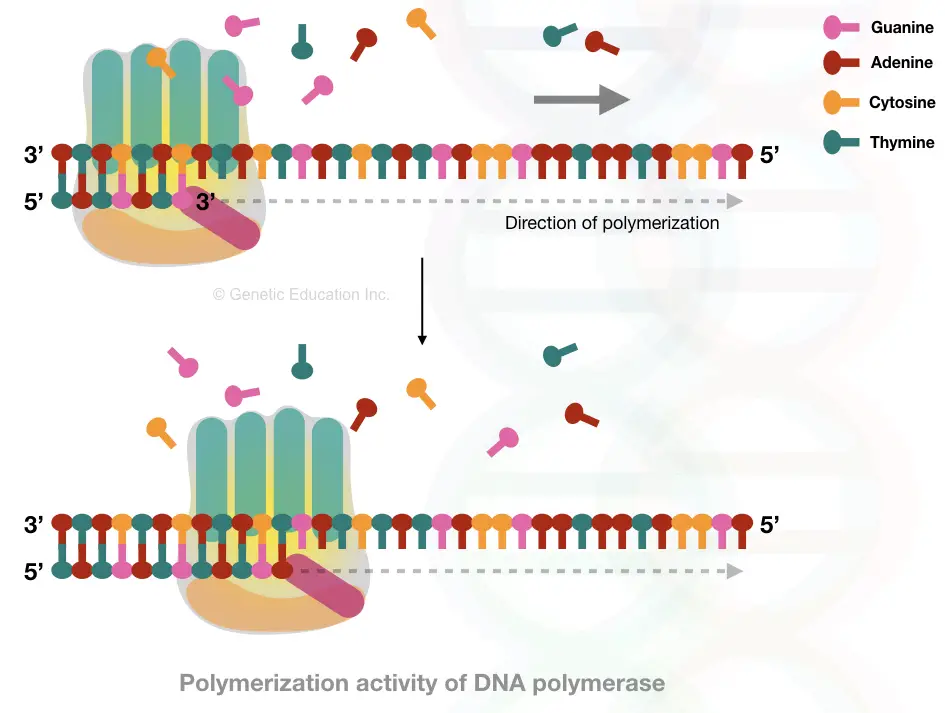
Elongation leads to the synthesis of a new DNA strand using the DNA polymerase. Enzyme DNA polymerase is a class of nucleic acid synthesis enzymes having a groove, when ssDNA passes through, a new nucleotide will be added, correctly.
However, elongation starts with a tricky thing, adding the short RNA primer. Similar to PCR- in vitro process of DNA synthesis, here also, a short single-stranded primer is required.
DNA polymerase can only incorporate nucleotides but it needs some starting point to replicate the substrate. RNA primase, a specialized enzyme synthesized a short RNA primer on the template DNA to settle the polymerase.
Once the DNA polymerase identifies the RNA primer, it settles on it, binds to the substrate- ssDNA and immediately starts polymerization. It starts synthesis from the 3’ open ends and adds exactly complementary nucleotides on the growing strand.
Once the process starts, primer is removed and polymerase fills the gap. See the image to learn more:
Leading and lagging strands
One strand of dsDNA is known as the leading strand while the other one is known as the lagging strand. You may wonder which one is which!
Both dsDNA is antiparallel, so the 3’ end from where polymerase starts working is our leading strand and the rest is the lagging strand. The synthesis process is a bit complicated, but I can understand, don’t worry about it.
Leading and lagging strand synthesis occur continuously and discontinuously, respectively.
Replication is initiated at the leading strand because the leading strand has a free 3’-OH end which is always recognizable by DNA polymerase. Here DNA polymerase runs toward the replication fork while on another strand (lagging strand) it runs away from the replication fork.
On the lagging strand DNA polymerase adds nucleotide away from the replication fork, meaning, it synthesizes DNA in fragments. Specialized sequences or fragments, called Okazaki fragments, present on lagging strands help it to promote synthesis in the exact direction.
Every Okazaki fragments have their RNA primer to initiate replication.
A primer in DNA replication
RNA primer starts the process of replication and is synthesized by the RNA primase enzyme. It is a short single-stranded RNA molecule; the ultimate need of DNA polymerase.
DNA polymerase uses the primer RNA and DNA junction as a substrate/ base to initiate replication. At a molecular level, the RNA primer provides a free 3’-OH end to the enzyme.
Until the DNA polymerase recognizes the 3’-OH end, it can’t start polymerization. The leading strand synthesis occurs in this manner. On the lagging strand instead of primase, the RNA primer is provided by the Okazaki fragment itself, because the process isn’t continuous.
Primase starts the synthesis of RNA when helicase initiates to unwind the dsDNA. In the newly formed duplex Note that during the process of PCR- an artificial DNA synthesis process, instead of RNA primer, a DNA primer set is used.
DNA polymerase progresses DNA replication from the 5’ to 3’ direction but can’t do it in 3’ to 5’ direction.
Termination:
Put simply, when all the DNA strands synthesize and DNA polymerase is removed, the process is terminated. Let me explain it to you in detail.
Many things are still left to complete. RNA primer should be removed, gaps between nucleotides should be filled, polymerase must be halted and proofreading must be completed. Note that Polymerase governs the process of proofreading.
We first need to understand replication, not on a single strand, but on a broad scale, consider all DNA of chromosomes. As we said, replication occurs simultaneously in many locations.
Thousands of replication forms are formed and DNA polymerase initiates replication in the opposite direction. So the enzyme itself is removed when it encounters another synthesized dsDNA.
But the gaps can’t be filled by the polymerase which is known as “nick”. Again, as replication initiates at thousands of locations so many nicks are formed and remain unfilled.
What fills the gaps will be discussed later, now jump to the RNA primer.
RNA primers that helped DNA polymerase to replicate must be removed as it isn’t a part of the genome. Proofreading removes RNA primer from the leading strand and each of the Okazaki fragments.
Another DNA polymerase fills the short gaps by adding “nucleotides” but still the gap can’t be filled between adjacent nucleotides, it can’t form the phosphodiester bond between adjacent nucleotides.
Those are “nicks” again.
Now after the end of all these, nicks remain unfilled in various locations. Here in the last step, another specialized protein came into function, known as “ligase”. It forms the phosphodiester bonds in gaps and fills nicks.
At the end of termination, completely continuously and exactly complementary daughter DNA will be formed.
The end replication problem and replication of telomere:
Everything isn’t perfect, and so is not the replication process. Polymerase synthesizes both strands but can’t complete replication all the time, meaning, when it comes to the end, several sequences can’t be duplicated.
On a linear chromosome, those sequences are our telomeres. Telomere is repetitive DNA, unusual sequences that are present on the ends and protect vital genes.
As we said, telomeric sequences can’t be replicated correctly and cause end replication problems all the time, over a period telomeres lose their strength.
Thanks to another type of polymerase known as “telomerase” it fills gaps after the end of replication. It indeed maintains the strength of the telomere. Telomerase has its own RNA primer to replicate.
Maintenance of replication:
In the background, many proteins and enzymes work to smoothly continue the replication process. The SSB protein- a single-stranded binding protein, binds to the single strand, immediately after helicase unwinds, to progress polymerase smoothly.
Studies suggest that in the absence of SSB, DNA will renature, and stop polymerase progression.
Another important protein is the sliding clamp. No one is talking about it, but it is important. The sliding clamp present on the base of DNA polymerase holds the enzyme and prevents slip-off.
The ring-shaped sliding-clamp helps in DNA polymerase progression until it completes the lagging strand synthesis. It helps slide polymerase efficiently on DNA strands without hurdles.
Topoisomerase I and II unwind the single-stranded and double-stranded DNA simultaneously and remove the tension on the rest of the DNA. Let me explain it to laymen, when DNA replication progresses, by unwinding, the remaining DNA gets twisted and ultimately stops replication.
Topoisomerase I and II cut and join DNA strands and remove tension by unfolding the twines.
Enzymes in DNA replication:
- DNA polymerase
Polymerase is an enzyme that synthesizes the new DNA strand by adding the nucleotides to the growing DNA strand. It recognizes special sequences present at the origin of replication and binds to those sequences.
Series of articles on DNA polymerase:
- Choosing the right DNA polymerase for your PCR experiment.
- Comparison Between: DNA Polymerase Vs RNA Polymerase.
- Helicase
Nucleotides on DNA strands bind with each other by hydrogen bonds. Helicase has specialized properties to unwind DNA stand by breaking the hydrogen bond between two strands. It creates a replication fork and two single strands.
Related article: What Is Helicase? And How It unwinds DNA?
- Topoisomerase
It is a group of special enzymes that releases the tension created on the rest of the DNA double helix. Imagine you unwind the rope by taking two threads of rope, what happened to the rest of the rope? Well, it will wind with each other, right?
A similar mechanism is applied here when DNA helicase unwinds the double helix, tension is increased on the rest of the DNA strand (supercoiling) which is removed by topoisomerase by cutting the double-stranded and single-stranded DNA by two different topoisomerase and further ligates DNA and removing the tension of strands.
- Primase
Primase synthesizes a short RNA primer for starting replication. Read more: Meet DNA Primase: The Initiator Of DNA Replication.
- DNA ligase
Ligase joins two DNA molecules. The gaps between Okazaki fragments and new DNA fragments are joined by ligase. Read more: What Is DNA Ligase? And How T4 DNA Ligase Works?
Wrapping up:
Replication is an essential process and occurs during cell division. There is a slight difference between prokaryotic and eukaryotic replication, however, the basic process and steps are the same. Eukaryotic organisms have a semi-conservative mode of replication.
The in vitro process of synthesizing the DNA is known as PCR- polymerase chain reaction which is a temperature-dependent enzyme governed process. It is used to study genes and DNA.
Initiation, elongation and termination are three important steps in replication, moreover, DNA polymerases have a major role in the process.
Sources:
Maffeo, C., Chou, H. Y., & Aksimentiev, A. (2019). Molecular Mechanisms of DNA Replication and Repair Machinery: Insights from Microscopic Simulations. Advanced theory and simulations, 2(5), 1800191. https://doi.org/10.1002/adts.201800191.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.



