A genomic library is a collection of clones/copies of smaller-sized genomic DNA fragments, used in various downstream applications.
Do you know?
- The human genome size is ~3.2 billion base pairs.
- The major part of the genome is non-functional or non-coding (97%).
- The first human genome sequence was completed in 13 years at the cost of 1 billion USD.
DNA library preparation is an important procedure in high throughput DNA sequencing, microarray and other hybridization-based techniques. Two types of libraries are cDNA libraries and genomic libraries prepared separately for different purposes.
Though the cDNA library-based techniques penetrated deep in genetic science and are popular & valuable; the genomic library, on the other hand, has unique credibility and applications.
Either type of library is a collection of fragments, largely; and are DNA fragments. But the question is why do we need fragments? Why do DNA libraries exist?
Our genome is so large. The unwind DNA strand of all cells can expand up to the sun, so it’s a difficult task to study a single DNA sequence, a part of a gene or a whole gene from the genome.
Technically, scientists have to face many problems, for example, non-complementary base pairing, reaction failure and so on. So the idea is what if we cut off our genome in small size fragments so that we can use a ‘pool of single types of fragments’ to investigate.
Put simply, investigating a 10Kb or 100Kb fragment is way more advantageous than a 3.2Bbp long genome!
Our genome is so huge, much like other eukaryotes where 97% DNA is nonfunctional, only 3% part has functional genes. Besides, It is structurally and functionally complex too. A portion considered as junk previously is now a hot research topic because it regulates gene expression.
Henceforth, evaluating the whole genome is imperative.
To study every possible sequence of our genome, we need it in fragments, accessible fragments, and that’s what our genomic library is. Once we prepare it, we can plan various experiments, determine hypotheses and explore new horizons of our genome.
So what exactly is the genomic library? Why is it needed and how to prepare a genomic library? All these questions and possible others, we will try to answer in this blog.
Stay tuned.
Key Topics:
What is the genomic library?
A genomic library is a fragmented DNA collection of the whole genome of an organism. The fragments are stored in bacterial or yeast vectors later on.
Properties of a genomic library:
- It should have all the genomic content of the genome of an organism.
- The size of fragments should be clone-able.
- The library represents the entire genome.
- The library should be screenable.
- Every fragment from the library should be processable enough.
Definition:
A clone/collection of similar types of genomic DNA fragments prepared by techniques like restriction digestion is referred to as a genomic library.
Genomic DNA library preparation:
This type of DNA library preparation process is simple, effective and less complex, unlike the mRNA library preparation. The entire process completes in 5 easy steps:
- Genomic DNA isolation
- DNA fragment generation
- Restriction digestion
- Mechanical shearing
- Cloning into a vector system
- Transforming into a host cell
- Library screening
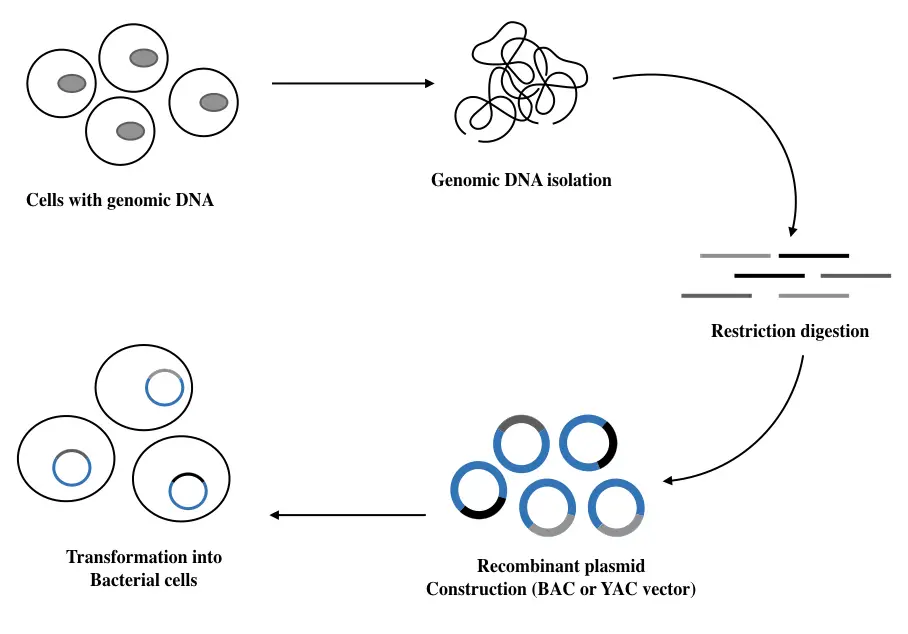
Genomic DNA isolation:
Genomic DNA isolation has been considered one of the easiest techniques in modern-day genetic science. There are a lot of standard protocols and ready-to-use DNA extraction kits are available on the market.
Keep in mind that the isolated genomic DNA should have purity around ~1.80 (at 260/280 absorbance) and quantity around or more than 100microgram.
You can read our previous article on this topic to understand the basics: Different types of DNA extraction methods.
Put simply, the isolation process includes steps of cell lysis, removal of protein and other impurities, DNA precipitation, DNA purification and DNA elution.
The extracted DNA can be run on 0.8% agarose gel to investigate its purity and quality manually.
DNA fragmentation:
DNA extraction is followed by DNA fragmentation where genomic DNA is parted into smaller random-sized fragments. As aforementioned, two common techniques for the purpose are restriction digestion and mechanical shearing.
But before going further into the article, you can read our previous article on this topic to get more clarity: DNA fragmentation technique.
Restriction digestion:
Restriction digestion is one of the most popular and widely used techniques in genomic library preparation. It’s a process governed by a restriction enzyme that cleaves off DNA into fragments.
The cuts are double-stranded, largely and can produce sticky or blunt ends. The enzyme binds to its recognition site, which is specific, unique and present throughout the genome many times. Take a look at the example below.
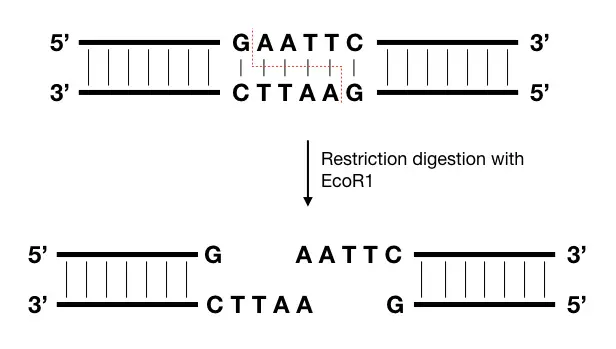
Sticky ends are more useful and easy to process than blunt ends, henceforth, the only restriction enzyme or endonuclease that can generate sticky ends are selected, for example, EcoR1.
Read more:
- What is Restriction Digestion and how to do it?
- What are the Restriction Enzymes? Top 10 Restriction Enzymes used in Genetic Engineering
Mechanical shearing:
A less popular but alternative method for fragmentation is mechanical shearing in which DNA fragmentation is done artificial and generates blunt ends. Mechanical processing needs sub-processing such as adaptor ligation and sticky end generation.
Here, a known linker or adaptor DNA is ligated to each DNA fragment and applied for restriction digestion to produce sticky ends.
Cloning into a suitable sized vector:
Now in the next step, processed fragments are incorporated into a suitable vector. In a layman, a vector is a vehicle that transfers the genetic material. Some common vectors used for genomic library preparation are plasmid, cosmid, phage, bacteriophage, lambda phage, BAC or YAC.
Note that vector preparation is an important step for achieving excellent end results. The vector is selected depending upon the size of the genomic DNA fragment. For example, to clone 150 to 200Mb DNA fragments, BAC- bacterial artificial chromosome is the best choice as it can carry 120 to 300 Mb DNA.
Here is the list of vectors, their size and DNA carrying capacity:
| Vector | Size | DNA carrying capacity in Kb |
| Plasmid | 1 to 200Kb | 15 |
| Phage lambda | 48.5Kb | 25 |
| cosmids | 35-50Kb | 45 |
| B. Phage | 35-100Kb | 70-100 |
| BAC | 100-200Kb | 120-300 |
| YAC | 100-3000Kb | 250-2000 |
After completion of the selection process, the vector is processed to incorporate DNA. To do so, the vector DNA which is usually a circular one! Is digestion with the same restriction enzyme as the genomic DNA.
A fragment and vector DNA is ligated using the ligase enzyme and sent for transformation.
Yet another crucial consideration in vector preparation is to use a selectable or screenable marker for validating insert. Screenable markers like the antibiotic gene, incorporated next to the fragment, facilitate the final screening process.
Transformation into host cells:
E.coli and yeast cells are two common organisms used as a host for genomic DNA transformation. The recombinant vector is inserted into the host cell to replicate there. It recombines with the bacterial or yeast chromosome and makes copies of vector genes and fragments of our genomic DNA.
Cells are cultured on agar media containing an antibiotic. So the cells having the antibiotic-resistant gene (our screenable marker) can only grow but not the rest. Meaning, only transformed host cells can grow.
Screening of genomic library:
Largely, two techniques applied for screening genomic fragment inserts are probe-hybridization and polymerase chain reaction. The probe-based hybridization technique is traditional, outdated and less accurate whereas the PCR technique is robust, speedy and more accurate.
In the probe-hybridization-based screening, a known probe sequence of 20 to 200bp, complementary to our genomic DNA fragment is allowed to hybridize and detected using autoradiography.
In the PCR-based amplification technique, a known primer set is allowed to amplify the insert, it also produces clones of that DNA. The untransformed plasmid DNA can’t amplify here.
So the accuracy of PCR is very high and at the same time, it produces additional copies of the genomic fragment too. After completion of the process, the amplicons are separated on agarose gel and stored for further investigations.
This is the simplest representation of how the genomic library is created and maintained. However, it consists of tedious experimental steps like cloning, culturing and transformation.
Advantages of the genomic library:
- The genomic library preparation process is comparatively simple and easy.
- A huge number of data can be generated.
- Fragments can be analyzed to assess SNPs and mutations.
- It is a comparatively speedy process.
- The fragment library can be stored for many years.
Disadvantages of the genomic library
- The size of individual fragments is too large hence sometimes sub-fragmentation is required for some downstream processing.
- Also, the larger fragment size is difficult to process and treat.
- The effect of a particular gene or protein can’t be studied.
Applications of Genomic library:
The genomic library preparation technique is widely used for years for DNA sequencing. It is nowadays also used for microarray analysis. Let us understand both scenarios with an example.
Genomic DNA library for DNA sequencing:
Genomic libraries allow scientists to sequence the whole genome using various techniques. Interestingly, the first genome sequenced by Sanger F is achieved by genomic library preparation.
In the process, fragments are re-fragmented by restriction digestion and applied for adapter ligation. Adaptors are known, oligonucleotide sequences, complementary to the primer set used for amplification.
Once every fragment is tagged with adaptors, are simultaneously amplified, sequenced and recorded. Later on, based on the location of the adapter, every sequenced DNA fragment is joined computationally to prepare a sequence of the entire genome.
Genomic DNA library for microarray analysis:
Let’s say we wish to investigate the polygenic nature of 15 different genes and other SNPs. We have all the probes prepared from the data of SNPs of each gene and immobilized on a chip.
Again the genomic DNA is sub-fragmented to increase the resolution of analysis and accuracy of results and are hybridized on a chip. Probes bind if they find complementation and give signals.
Signals are recorded for each probe, are evaluated at the end of the experiment.
Moreover, on a broader note, genomic library preparation has applications in SNP analysis, mutational detection, copy number variation studies and population-based genetic studies.
Wrapping up:
Much like the cDNA library preparation, the genomic library has crucial importance in molecular genetic analysis. In recent times, it has great utility in polygenic disease analysis and investigating thousands of SNPs in a single assay.
However, the process is a bit time-consuming and costlier, it also needed manpower too. Genomes of many different organisms are prepared through the genomic library.


