“Inverse PCR is used for site-directed mutagenesis and insert studies. Learn about the technique, concept, protocol and applications of inverse PCR.”
PCR (Polymerase Chain Reaction) is a technique to amplify and study a DNA sequence. However, using a conventional PCR variant for every application is difficult. So scientists have to modify the basic approach for expanding its usability.
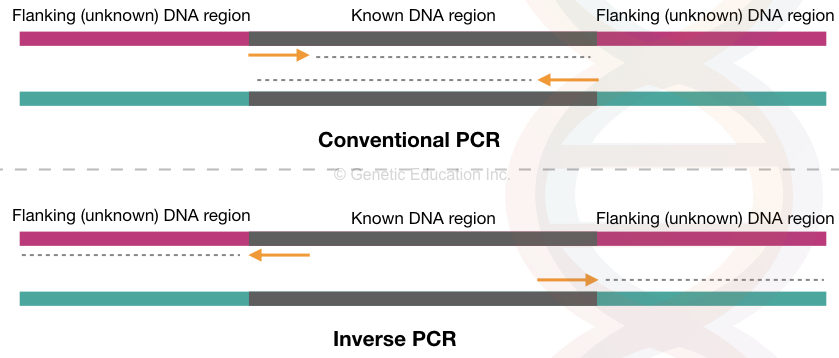
Such modifications increase the specificity, sensitivity and detection range. One such modification is inverse PCR. Conventional PCR way can amplify the known region inward using the primer set. We know it as the target region.
But what if we want to amplify the regions flanking our target sequence or the sequence opposite to it? That’s possible using the inverse PCR approach.
Sounds confusing! No worries.
In this article, I will explain to you the concept of inverse PCR, its principle, procedure, protocol, applications and limitations.
Stay tuned.
Disclaimer: Information provided in this article is collected from peer-reviewed resources and re-presented in an understandable language. All the sources are enlisted at the end of the article.
Key Topics:
What is Inverse PCR?
Conventional PCR amplifies the target DNA sequence or a gene in a cyclic-enzymatic reaction using a set of primers. The forward and reverse primers synthesize the target location inward.
Here, the target sequence is known to us. Using the available information we design primers for amplification. However, any flanking or outside sequence information can not be obtained this way.
Any insertion or transposition in the flanking sequence can only be studied if we can amplify them. In the IPCR or inverse PCR, the target sequence-specific primers are designed in an inverse position.
So, instead of amplifying inward, it amplifies outward/flanking sequence. The sequence upstream, downstream or both to the target can be amplified and studied in this way.
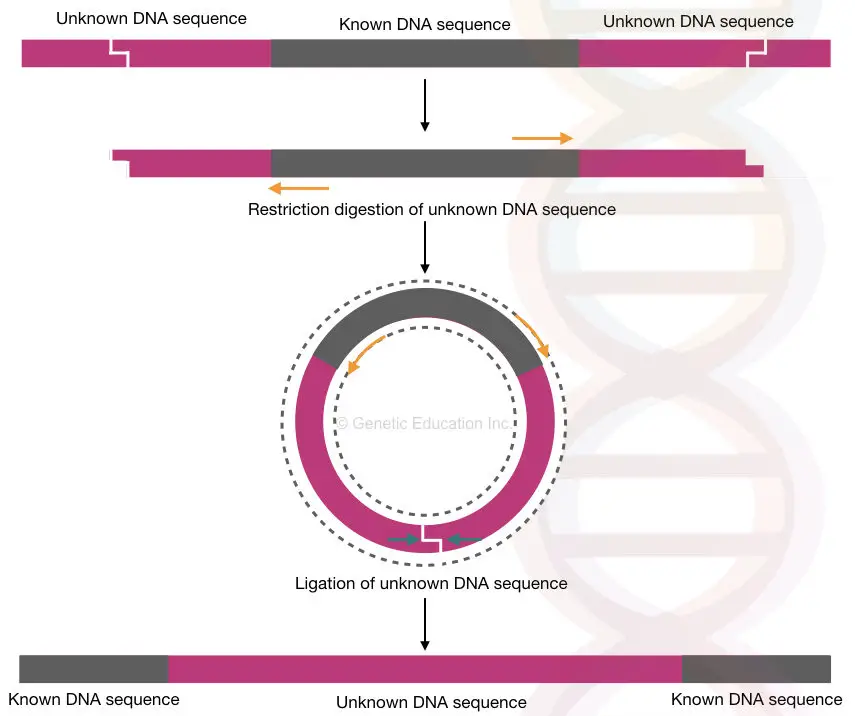
However, our genomic flanking region can be of any length and can not be effectively amplified. So what scientists do is, they first digest the flanking region with a restriction enzyme, then ligate the ends, circularize it and amplify it.
Sounds good!
Now, for detection and analysis, either conventional gel electrophoresis or DNA sequencing can be performed. The method was conceptualized by Ochman H et al. (1988).
Keep in mind that no specific requirement is needed. The amplification reaction is completed using the standard ingredients i.e. dNTPs, Taq DNA polymerase, water, buffer, primers and template DNA. Other names of iPCR are inverted PCR and inside-out PCR.
iPCR is employed to study unknown genomic regions, transposition sites, viral insertion sites, genome walking and chromosomal rearrangements. Furthermore, it is used in cloning and genetic engineering experiments.
Principle of Inverse PCR
Inverse PCR is a method used to amplify the flanking regions adjacent to a known DNA sequence or an inserted DNA fragment. The primers are designed in a way that amplifies outward to the target sequence.
Restriction digestion is applied to the genomic DNA to selectively cleave the flanking regions, leaving the known DNA sequence intact. The cleaved flanking regions are then ligated together to form circular DNA molecules, which are amplified using the same primers.
Steps and Procedure
- Target selection
- DNA extraction
- Restriction digestion of flanking sequences
- Circularization using ligation.
- Amplification of ligated circular DNA.
- Detection
Let’s understand each step one by one.
Target selection:
Succession of the whole experiment relies on the selection of the target. Although we want to study an unknown flanking region, here our target is the known DNA sequence. Using the known sequence information the primers have been designed in a reverse position.
Keep in mind that the target known sequence should not contain the restriction site for the selected enzyme. Also, the unknown flanking region should not flank more than 2Kb. The basic PCR has a limited amplification power.
Related article: How to Find Information about a Gene of Interest?

DNA extraction:
Using an available protocol or ready-to-use kit the genomic DNA is extracted from the sample. As usual, the quality and quantity have been assessed before using the DNA further. These are our articles on DNA extraction. You can read it.
- Different types of DNA extraction methods
- Phenol-chloroform DNA extraction method
- CTAB DNA extraction method
- Proteinase K DNA extraction method
Restriction Digestion:
Unlike the routine amplification methods, in inverse PCR, the experiment begins with restriction digestion. Using a selected restriction enzyme (which can not cleave anywhere in the target/known sequence) the genomic DNA is cut.
REases can cut the flanking as well as other genomic DNA having the recognition sites. Fragment separation and validation have been done on the agarose gel electrophoresis. In the case of eukaryotic DNA, thousands of fragments can be obtained.

Circularization using ligation:
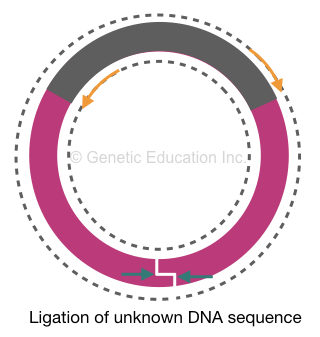
Before amplification, the restriction digestion genomic DNA fragments are circularized using a ligation reaction. The fragment mixture is treated using a ligase enzyme that reseals two ends of each fragment and makes them circular.
PCR amplification:
Now, here comes the interesting part.
Our target sequence using which we have prepared PCR primers is circularized and intact. But the flanking region is digested and circularized. Here, the primers will bind to their complementary location on the target sequence.
As per our plan, instead of amplifying the target DNA, it will expand outward and amplify the flanking region. The amplification of whole circular DNA will generate two single-stranded linear amplicons for another round of amplification.

Analysis and Detection:
Conventional gel electrophoresis or DNA sequencing can be used for detection and analysis. Noteworthy, the gel electrophoresis can give us an idea about length polymorphism and amplification succession.
It can not provide sequence-related information. Whereas, sequencing provides information on DNA sequence. Using bioinformatics tools, the sequence can be analyzed and studied. Any insertion or transposition in the flanking region can be studied this way.
Inverse PCR Protocol
Select the target and design the primers using online tools. Don’t know how to design PCR primers? Click the link and read the article.
Extract DNA using phenol, chloroform and isoamyl alcohol DNA extraction method. Click the link for detailed protocol. You can also use any other protocol as well. Also, this guide will help you choose the right DNA extraction protocol.
Select a restriction enzyme. (Demonstration guide: How to choose a restriction enzyme).
Use 5µG genomic DNA for restriction digestion. Perform digestion reaction at 37ºC for 12 hours. Follow this table for reaction preparation.
| Reagent | Concentration |
| gDNA | 5μl (Total 5μg) |
| RE buffer (5X)* | 4μl |
| REase (restriction endonuclease) (1 unit) | 2μl |
| D/W | 9μl |
| Final volume | 20μl |
*If the concentration of the RE buffer is 10X use only 2μl of buffer for the reaction.
*Detailed protocol is provided by the kit supplier.
Inactive the enzyme at 65ºC for 20 minutes to stop the digestion reaction.
Run the fragments on 3% agarose gel. (Demonstration guide is here: Agarose gel electrophoresis protocol).
Re-extract the DNA with a standard protocol or ready-to-use kit.
Perform ligation using 100nG to 1µG purified DNA. Refer to the present table for ligation reaction preparation.
| Reagent | Quantity |
| Template DNA | 100ng to 500ng |
| Ligase enzyme | 5 μl ( 1 unit or 1μl for 5 units) |
| Ligation buffer | 10μl |
| ATP (optional) | 10μl (10mM) |
| DD/W | Final volume 100ml |
*Detailed protocol is provided by the kit supplier.
Use T4 (Bacteriophage) DNA ligase having optimum activity at 16ºC. Incubate the reaction at this temperature overnight.
Again, re-extract the circularized DNA using a standard protocol or spin-column DNA extraction kit. This step will remove all the unligated and non-circularised DNA fragments.
Dissolve the DNA in the TE buffer. (The TE buffer preparation guide is here click the link).
Now prepare the PCR reaction using the PCR reaction preparation protocol given in the table below.
| Component | Stock | Concentration | Quantity |
| dNTPs | 200µM | 200µM each | 4µL |
| PCR reaction bufferWith MgCl2 | 10X | 2X | 5µL |
| Taq DNA polymerase | 5U | 1U | 0.5µL |
| Forward primer | 100pM | 200pM | 2µL |
| Reverse primer | 100pM | 200pM | 2µL |
| Ligated DNA | 250 ng | 500 ng | 2µL |
| Water | 9.5µL | ||
| Total | ————————– | 25µL |
>> Check out this demonstration for PCR reaction preparation.
Place all the reaction tubes in the PCR machine and set the protocol as given in the table below.
| PCR Steps | Initial Denaturation | Denaturation | Annealing | Extension | Final extension |
| Temperature | 90 ̊C-95 ̊C | 90 ̊C-95 ̊C | 55 ̊C-60 ̊C | 72 ̊C | 72 ̊C |
| Time | 5min | 50 sec | 50sec | 2min | 7 min |
| ——————– | ——————- | 25-30 cycles | ————— | ——————— |
Upon run completion, validate the PCR amplicon on 2% agarose gel.
After that send the amplicon for DNA sequencing and analysis.
Applications of Inverse PCR
Now, let’s understand why we need to use inverse PCR and when it is useful.
Identification of a flanking region:
One of the pivotal and widespread applications of the iPCR is to study and characterize the gene flanking region. Amplifying and studying the flanking region gives us information regarding the gene promoters or enhancers.
Transposon studies:
Another crucial application of the present technique is in the transposon and jumping gene studies. Scientists can investigate and identify transposon insertion sites (upstream or downstream) near a gene.
Such studies help in the genotype-phenotype correlation and understand the impact of transposition on gene expression. Further, it’s also used in transposon tagging experiments and other related studies.
Detection of chromosomal rearrangements:
Yet another fascinating approach is in studying the chromosomal rearrangements such as insertion, deletions, and duplications by amplifying the rearrangement spanning regions. In addition, gene fusion and oncogenic rearrangements are studied. Such analysis can be helpful in disease and cancer research.
Insert validation:
iPCR is also employed for the detection of transgene, plasmid DNA, viral gene segment, and viral vector insertion in the genome. Through the amplification of a flanking region near the target DNA, scientists can investigate and validate the insertion.
Genetic engineering:
One of the pivotal applications of the iPCR is in gene cloning and genetic engineering experiments to validate insertion in the host genome.
Related article: Genetic Engineering: Importance and Educational Requirements.
Site-directed mutagenesis:
The present technique is also used in site-directed mutagenesis studies. Here the mutation insertion can be validated using the inverse PCR approach. By doing so scientists can investigate the outcome of mutagenesis.
Limitations
Despite having amazing applications in various genetic fields, inverse PCR has several serious limitations as well.
- The PCR has limited amplification capacity. A normal PCR protocol can amplify 1.5 to 2 kb DNA fragments effectively. Thus, flanking regions within this range are amplified.
- The involvement of enzymatic steps, repetitive extraction and reaction preparation makes it a time-consuming, complex and costly method.
- As the reaction involves many enzymatic steps and incubation, there are high chances of reaction failure.
- One of the major limitations of the present method is the requirement of prior knowledge for at least one sequence adjacent to the flanking target.
- It can not be employed on novel and less-characterized genomic regions.
Wrapping up:
In conclusion, iPCR can not be used for routine research and clinical diagnosis due to its complex and tedious nature. However, its utility in mutagenesis studies and insert validation is so important.
It’s also important to note that quantitative iPCR can help us quantify the amount of flanking or insert present near the target sequence. Such applications make inverse PCR an important technique in genetic research, particularly in genetic engineering.
I hope you like this article. Do share it and subscribe to our blog.
Resources:
Ochman, H., Ajioka, J.W., Garza, D., Hartl, D.L. (1989). Inverse Polymerase Chain Reaction. In: Erlich, H.A. (eds) PCR Technology. Palgrave Macmillan, London. https://doi.org/10.1007/978-1-349-20235-5_10
Silva D, Santos G, Barroca M, Collins T. Inverse PCR for Point Mutation Introduction. Methods Mol Biol. 2017;1620:87-100. doi: 10.1007/978-1-4939-7060-5_5. PMID: 28540701.
Green MR, Sambrook J. Inverse Polymerase Chain Reaction (PCR). Cold Spring Harb Protoc. 2019 Feb 1;2019(2). doi: 10.1101/pdb.prot095166. PMID: 30710023.
Huang SH. Inverse polymerase chain reaction. An efficient approach to cloning cDNA ends. Mol Biotechnol. 1994 Aug;2(1):15-22. doi: 10.1007/BF02789286. PMID: 7866865.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.