“The pre-existing, newer and futuristic genetic technologies revolutionized medicine by advancing healthcare diagnosis and clinical research. Explore their applications in this article.”
We are now in the era of having the best genetic technology advancements in terms of throughput, precision, TAT and cost. New-age technologies like NGS and CRISPR, futuristic opportunities like SMRT and digital PCR and of course the old-school approaches like PCR and Sanger sequencing have been extensively instrumental in medicine.
These technologies under a single roof, revolutionized medicine and clinical research, diagnosis and treatment.
I visited many genetic facilities in the last 6 months and understand the importance of these technologies in the context of the medical and healthcare industries. Trust me, it changed the present field, entirely.
So I came up with an editorial review article from Genetic Education to show you the immense and diverse proportion of how genetic technologies are transforming our medical facilities and revamping the meaning of disease research, diagnosis and treatment.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy to understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Key Topics:
Genetic Technologies
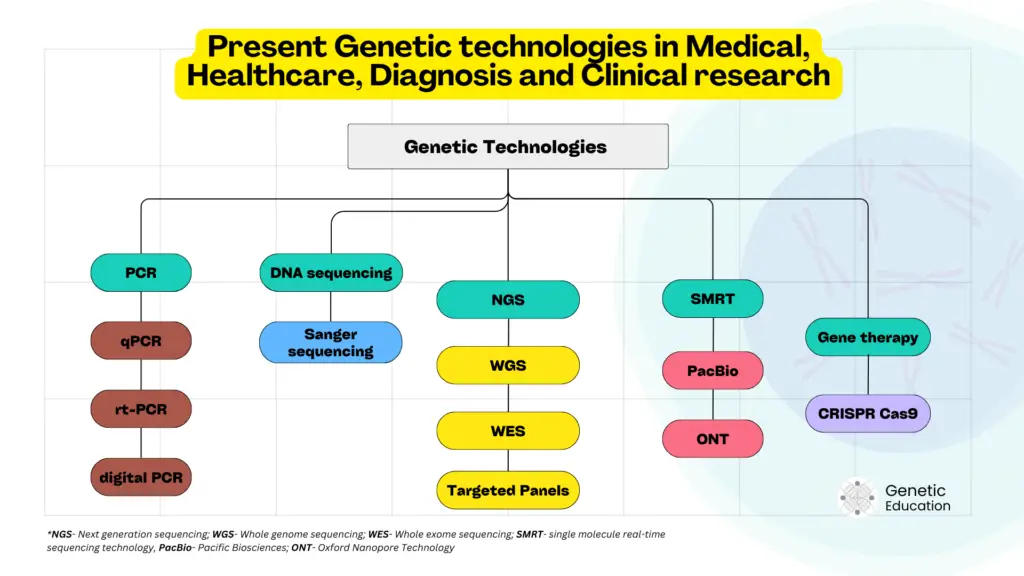
Now, we will categorize all the technologies and their advanced version we have, in three different categories,
- The old-school approaches like Karyotyping, FISH, PCR and Sanger sequencing
- The newer NGS and microarray-based approaches
- The Futuristic SMRT technologies
- Gene therapies and CRISPR-cas9
PCR technologies
PCR (Polymerase Chain Reaction) has been instrumental in clinical and medical setup for many years. It is useful particularly to investigate monogenic and inherited conditions. However, the advanced versions are available now, making it a better option for the same field.
Besides, its conventional applications, reverse transcription and quantitative PCR have been extensively and routinely used in diagnostics to investigate gene expression and identify and quantify pathogens, and infections.
For instance, quantitative PCR is a fast, easy and effective diagnostic option for COVID-19, Tuberculosis and HIV. Furthermore, cancer-causing gene variants can be quantified using the same approach.
Interestingly this allows doctors to understand the infection progress and cancer stage. The futuristic approach— the droplet digital PCR, the latest innovation in the PCR — allows even more accurate analysis of rare gene variants and more precise expression analysis by partitioning each reaction into smaller-sized droplets.
The digital PCR is capable enough to find out mutations from low abundance DNA samples such as circulating tumor DNA with high precision.
Despite having low throughput, the requirement of prior sequence information and extensive sample preparation, PCR technology is a cornerstone for medicine, diagnostics, healthcare and clinical research.
DNA sequencing technologies
The stand-alone and only DNA sequencing technique we have is the Sanger sequencing. Automation made it more powerful, trustworthy and handy, though. It can investigate what the PCR can do! But at a sequence level.
DNA sequencing allows one to read the nucleotide sequence of a DNA or gene. This can identify gene variants associated with a disease, inherited condition, or even cancer and identify pathogens from the sample.
In diagnostics, the present technology enables doctors to identify genetic mutations associated with an inherited condition thereby guiding targeted therapies. For instance, detection of BRCA1/2 gene mutations.
DNA sequencing has been instrumental in medicine, diagnostics and clinical research for more than two decades and will still remain relevant in upcoming years.
NGS- technologies
After the introduction of the massively parallel NGS (Next-generation sequencing), the entire medicine, diagnostics, healthcare and clinical research industry has been revolutionized.
It has overcome the limitations of the traditional Sanger sequencing technique by providing genome-wide sequencing coverage.
Techniques like whole-genome and whole-exome sequencing allow determining disease-causing variants from the entire genome and coding regions, respectively, though whole-exome sequencing has more advantages in diagnostics.
The main USP of NGS, the throughput makes it a standout technology in comparison to other genetic technologies and therefore used not only in diagnostics but also in clinical research.
Cancer, cardiomyopathies, neuroscience, reproductive medicines and infertility, prenatal and postnatal, newborn screening, and other medicine fields now extensively rely on the NGS.
NGS also investigates rare genomic variants more effectively through its massive parallel sequencing technology and high sequencing depth. Hence, rare somatic and sporadic cancer variants can be identified and linked with the cancer.
The development of novel gene panels for a particular condition and disease made it even more accessible, handy and accurate for clinical diagnosis. Medical professionals routinely use such gene panels for the diagnosis of complex diseases.
Microarray and gene expression technology
The old-school techniques like karyotyping and a low through FISH, lack genome-wide coverage due to low resolution. The most advanced version of chromosomal analysis, the array CGH or chromosomal microarray technology filled this gap.
Microarray technology can investigate thousands to millions of copy number variations from all 23 pairs of chromosomes without requiring cell culture or fluorescence hybridization. Not only that, but it can even determine gene expression using gene expression microarray assay.
Microarray is the most trusted, widely used and nearly a gold-standard technology now in medicine and healthcare, diagnosis and clinical research.
It can investigate expressions of low genomic variants, classify cancer based on the subtypes, identify drug responses and help in personalized treatment and medicine. The ready-to-use panels make it even more useful for the same field.
SMRT technology
The futuristic SMRT technology, which is single molecular real-time sequencing will change the whole sequencing industry. This technology works on the simple principle in which a single molecule helps to read the nucleotides when the single-stranded DNA passes through the membrane.
Each nucleotide is determined and read in real-time here. An additional long-read sequencing approach makes the technology even more powerful and way ahead of other previous-generation NGS platforms.
PacBio and Oxford Nanopore Technologies offer SMRT sequencing that is capable enough to perform all the applications the NGS can do. The present technology is still not widely available in medicine and healthcare due to its low accuracy.
Gene therapies and Gene editing technology
The most promising genetic technologies that can help treat genetic diseases are gene therapies and gene editing. The main goal here is to repair or replace the fault and mutant gene from the patient and replace it with a healthy one.
Using either non-viral or viral vectors, the therapeutic gene is introduced into the affected or patient’s cells. However, each approach has its own advantages and limitations.
Non-viral vectors are safer but least effective, whereas viral vectors are more effective but unsafe to use. However, FDA-approach gene therapies for muscular dystrophy and other untreated conditions are now available.
Research is ongoing to develop novel gene therapy approaches for different genetic conditions.
CRISPR-cas9 technology
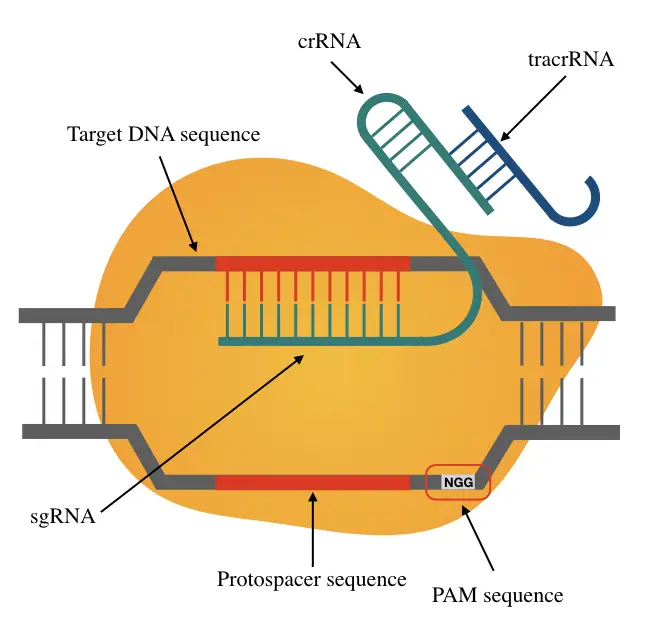
CRISPR cas9 technology has revolutionized gene editing technology with its immense potential and ability to accurately edit genes. CRISPR can not only remove fault genes but can also repair mutations, deactivate disease-causing genes or even accurately incorporate a therapeutic gene.
Using the cas9 protein the CRISPR sequence (Clustered Regularly Interspaced Short Palindromic Repeats) identifies the target region, attaches to it and performs the modification.
Clinical trials showed promising results and many therapies are available soon. CRISPR-cas9 is the future of medical treatment and the healthcare industry.
Wrapping up:
These are the genetic technologies that are used in medicine, healthcare, diagnosis and clinical research routinely. Notedly, NGS is now widely used in the clinical field due to its high precision, throughput and wide coverage.
Contrary, gene therapies aren’t still available for full use. However, in the near future, thousands of gene therapy treatments will be available for many genetic diseases.
I hope you like this article. Do share and subscribe to Genetic Education.
Resources:
- Lynch, J. R., & Brown, J. M. (1990). The polymerase chain reaction: current and future clinical applications. Journal of Medical Genetics, 27(1), 2–7. https://doi.org/10.1136/jmg.27.1.2.
- Nazir, S. (2023). Medical diagnostic value of digital PCR (dPCR): A systematic review. Biomedical Engineering Advances, 6, 100092–100092. https://doi.org/10.1016/j.bea.2023.100092.
- Seifi M, Ghasemi A, Raeisi S, et al. Application of Next-generation Sequencing in Clinical Molecular Diagnostics. Brazilian Archives of Biology and Technology. 2017;60. doi:https://doi.org/10.1590/1678-4324-2017160414.
- Ardui S, Ameur A, Vermeesch JR, Hestand MS. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018;46(5):2159-2168. doi:10.1093/nar/gky066.Trevino, V., Falciani, F. & Barrera-Saldaña, H.A. DNA Microarrays: a Powerful Genomic Tool for Biomedical and Clinical Research. Mol Med 13, 527–541 (2007). https://doi.org/10.2119/2006-00107.Trevino.
- Li T, Yang Y, Qi H, et al. CRISPR/Cas9 therapeutics: Progress and Prospects. Signal Transduction and Targeted Therapy. 2023;8(1):1-23. doi:https://doi.org/10.1038/s41392-023-01309-7