“Explore the differences between single-end and paired-end sequencing and their advantages and limitations in this article.”
Next-generation sequencing platform relies on the principle of DNA fragmentation. A DNA sample is fragmented into smaller fragments and read in the machine by sequencing by synthesis or other chemistry (depending on the instrument).
Individual fragments are read either from the single end or from both ends, named single-read sequencing and paired-end sequencing, respectively.
Single-end and paired-end sequencing, both are the NGS technology advancements by Illumina. Developed for Next-generation sequencing for DNA and RNA, single-end and paired-end sequencing has been known for their capacity to sequence the whole genome.
However, both are substantially different and utilized for different applications. In this article, we will understand the differences between the single-end and paired-end sequencing.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Single-End vs Paired-End Sequencing
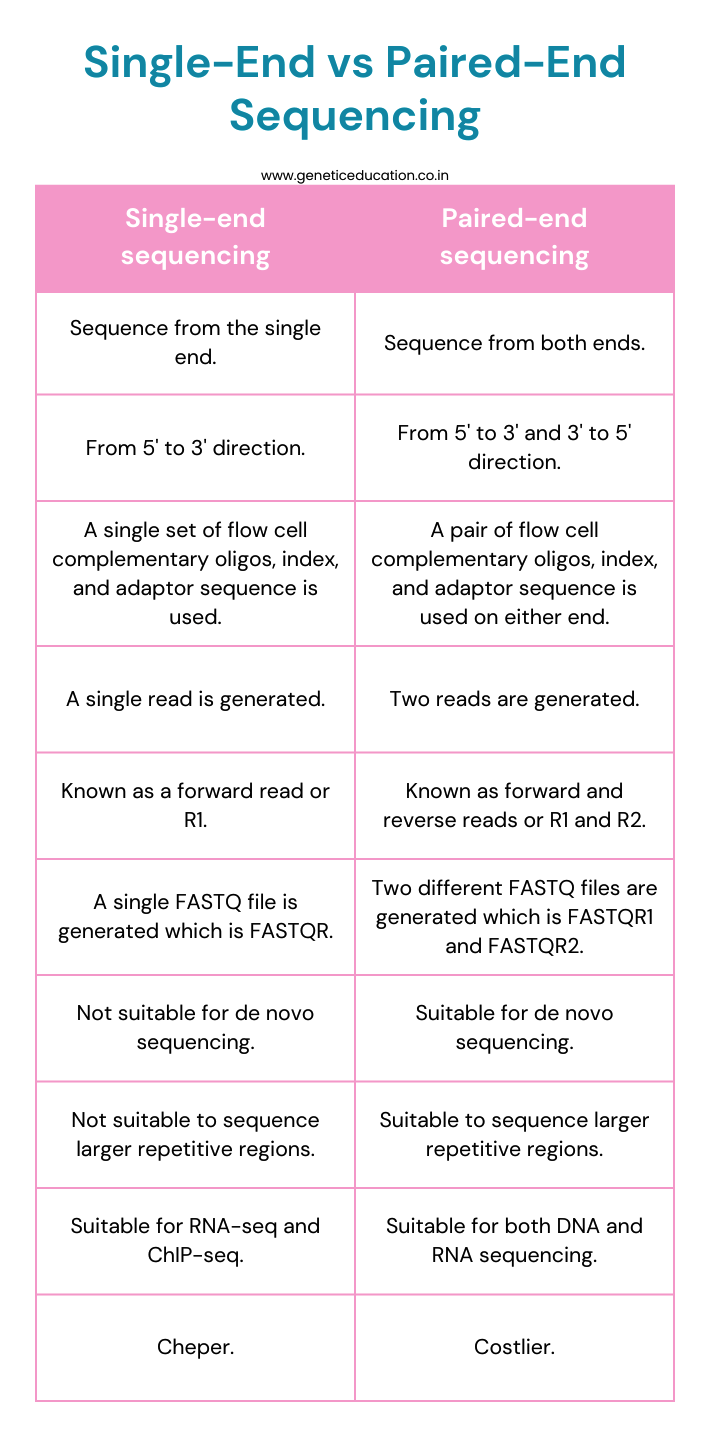
In single-end sequencing, the sequence is read from only a single side, usually from a 5’ to 3’ direction while in paired-end sequencing, the sequence is read from both ends, from a 5’ to 3’ and 3’ to 5’ direction.
Note that both work on the principle of sequencing by synthesis mechanism in which each nucleotide is added to the sequencing strand and read (using a colored tag) at the same time.
In single-end sequencing, a single set of a flow complementary sequence, index sequence and adaptor sequence has been used, while in paired-end sequencing two sets of flow complementary sequence, index sequence and adaptor sequence have been used.
Thus, in paired-end sequencing, it is named P1 and P2, index 1 and index 2, and adaptor 1 and adaptor 2. Each set is attached on either side of a target sequence.
Single-end sequencing only generates a single read which is read 1 or R2 and hence, only a single FASTQ file. Conversely, paired-end sequencing generates two separate reads– R1 and R2 and hence, two separate FASTQ files, FASTQR1 for the first or forward read and FASTQR2 for the second or reverse read.
The flow cell, which is a microscopic slide on the Illumina NGS platform contains a single type of hybridization flow cell oligonucleotide sequence for single-end sequencing and two different types of flow cell oligonucleotide sequences for paired-end sequencing.
Thus, during single-end sequencing, the amplification that occurs on a flow cell is linear while paired-end sequencing allows bridge amplification.
In the bridge amplification, one end of the target sequencing hybridizes with the flow cell oligonucleotide sequence 1 while the other end with sequence 2, forms the bridge for amplification.
Paired-end sequencing provides high-quality alignment and sequencing of larger repetitive regions whereas, single-end sequencing can’t effectively sequence such genomic loci.
Paired-end sequencing further can perform de novo sequencing by preparing long contings, contrary, single-end sequencing can’t prepare long contings and thus is not suited for de novo sequencing.
Paired-end sequencing generates twice the number of reads for the same sequence in comparison to single-end sequencing. Meaning, that the more the number of reads, the more the accuracy of sequencing.
Thus, paired-end sequencing is the most accurate and precise NGS optimization compared to single-end sequencing.
Single-end sequencing can’t effectively study genomic insertions and deletions, as the sequence is read from both sides during the paired-end sequencing, such genomic alterations can be effectively studied.
Despite having these advantages, single-end sequencing is cheaper and cost-effective while paired-end sequencing is costly as it utilizes additional reagents and computational power.
Applications:
| Single-end sequencing | Paired-end sequencing |
| Whole genome, whole exome and whole transcriptomics studies. Not suitable for de novo sequencing. Suitable for RNA-seq and ChIP-seq. | Whole genome, whole exome and whole transcriptomics studies. De novo sequencing. Study repetitive DNA regions. Genetic variations such as indels and gene fusions. |
Related recent articles:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- What is NGS?- Definition, Principle, Steps, Chemistries, Advantages and Limitations
- De Novo Sequencing: Steps, Procedure, Advantages, Limitations and Applications
Wrapping up:
Both, single-end and paired-end sequencing work on ‘sequencing by synthesis’ chemistry and are the best NGS platforms, however, due to additional benefits of paired-end sequencing, it has been utilized in cancer and gene expression studies.
Still, scientists are using single-end sequencing in many cases to reduce the cost, for example, chromatin immunoprecipitation sequencing.
If you are a research student or want to start up a genomic testing facility, the present comparison would definitely help in making the decision. I hope you like this article. Please share this article in your circle or on social media.
Sources:
Advantages of paired and single-read sequencing by Illumina.


