“Reversible Terminators used in NGS allow incorporation of a single nucleotide at a time thereby resolving complex genomic sequencing challenges. Learn about reversible terminators in this article.”
Sequencing by synthesis is an amazing NGS chemistry and a successful one! Almost every second generation of short-read sequencing platforms uses the SBS approach, majorly. Although, use different detection methods.
For instance, the Illumina sequencing platform uses a fluorescence (4-, 2- and 1-channel) detection approach while Ion Torrent uses Hydrogen Ion detection using a semiconductor approach. But the heart of each detection method is SBS.
In this sequencing approach, the sequencing occurs just like normal DNA replication. The polymerase adds nucleotides to the template DNA strand and each nucleotide is detected using a platform-specific detection method.
However, the present approach can not sequence entire genomic content, effectively, particularly the homopolymeric regions. Optimizing the SBS chemistry using a reversible terminator by platforms such as Illumina/Solexa solved the present problem.
In this article, I will explain to you what the reversible terminator is and why it is needed in the NGS.
Stay tuned.
Key Topics:
What is a Reversible Terminator?
A reversible terminator is a modified nucleotide base analog (A, T, G and C-analog) used during the SBS chemistry to add “one nucleotide at a time.” The present sentence is very crucial. We will discuss it in the later part of this article.
The first-generation Sanger sequencing techniques work on the principle of chain termination. When the polymerase encounters the modified ddNTP, it terminates the chain synthesis, permanently.
Whereas, in the SBS- reversible termination technique, the chain synthesis or we can say the primer extension is terminated temporarily until the sequencing signal is detected. After that, the blocker or terminator is removed and allows the incorporation of another nucleotide (analog).
This is a substantial difference between the first-generation chain termination technology and next-generation reversible chain termination technology. The reversible terminator approach was introduced by Dr Jingyue Ju from Colombia University.
If you want to know about various sequencing generations, read this article: What is First, Second and Third Generation Sequencing?
Mechanism of Reversible termination
We will not discuss all the SBS chemistry steps, as we wrote a dedicated article on this topic. We will start with immobilization.
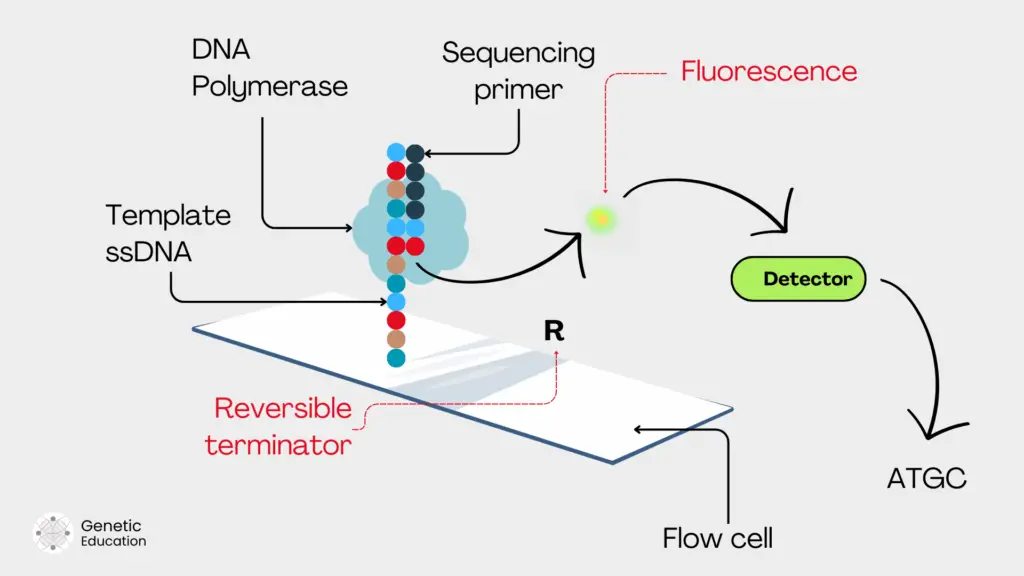
Immobilizing on solid support:
So first our libraries are immobilized along with the sequencing primers on the solid support. For instance, a flow cell in the case of Illumina. Now the flow cell already consists of the adaptor complementary sequences to template hybridization.
Note that the flow cell also contains a ‘specific’ polymerase to regulate the SBS process.
Primer extension:
Now, in the next step, the polymerase came into the picture and started the catalytic reaction using the free 3’ OH end of the primer. It incorporates the first nucleotide (which is our nucleotide analog with a reversible terminator).
It extends the primer at one nucleotide addition. The nucleotide (analog) contains a fluorochrome and a blocker or terminator molecule, for detection and termination, respectively.
The terminator is present probably on the 3’ OH end and blocks further extension. The polymerase will remain there until the detection is completed.
Detection:
Before detection, the first round of washing is conducted. This will remove all the unincorporated and unused nucleotides to avoid signal disruption. Now, the fluorophore is released from the nucleotide.
Using a laser light the fluorescent is emitted. The high-definition camera captures the image and is detected by the detector. Meanwhile, the terminator still holds down the primer extension.
Removal of fluorescent tag and terminator:
Immediately after fluorescent detection, the terminator or blocker molecule is removed by washing from the nucleotide. Now, the 3’ OH end is ready for another round of synthesis.
This allows further primer extension. And the process occurs in the cycle for all four nucleotide incorporation. Lastly, all the components are washed off from the flow cell before another round of nucleotide incorporation.
Principle of Reversible Terminator:
The modified nucleotide analog (fluorophore + Reversible terminator) when added at a complementary location on the template DNA, temporarily blocks the primer extension until the fluorescent signal for the nucleotide incorporation is detected in SBS chemistry.
After detection, the fluorophore and blocker are removed and washed off from the flow cell to allow the second round of nucleotide incorporation.
Why is the reversible terminator used in NGS?
Adding the reversible terminator approach in the sequencing chemistry increases the sequencing time as well as complicates the process, so why do we do this?
Homopolymeric regions are the darker genomic regions where a single nucleotide is present repeatedly more than two times. For instance, AAAA, TTTT, GGGGGG and CCC. Thus, such regions are also known as mononucleotide microsatellites.
Previous generation sequencing approaches, for instance, pyrosequencing (the first high throughput sequencer) face challenges in sequencing such regions. Pyrosequencing generates a pyrogram for each nucleotide addition.
The peak height in the pyrogram or electropherogram is directly proportional to the amount of nucleotide addition. If two consecutive nucleotides are added then, there are twice the height of the peak and so on.
So when it encounters a homopolymeric region, it can not convert the signals effectively into the peaks or output because all the signals are detected as a single concentrated signal unit as there is no blocker for each round of nucleotide addition.
This restricts its use in whole genome sequencing. Even in the chain termination approach (Sanger sequencing) the presence of homopolymeric sequences results in non-specific and poor-quality sequencing due to the ineffective function of an enzyme.
To overcome these challenges the reversible terminator is incorporated in the SBS chemistry. Now, you understand why I asked to note down “one nucleotide at a time?”
This is the reason.
If you have a question about what if the reversible terminator is not added in the NGS chemistry, you have the answer now! The homopolymeric region can not be effectively sequenced. This will decrease the accuracy, sensitivity and efficiency of NGS.
Related article: 10 Challenges in Whole Genome Sequencing.
Types of reversible terminators
Various approaches are developed to use the reversible terminator approach for various platforms. Here is the comprehension of each type.
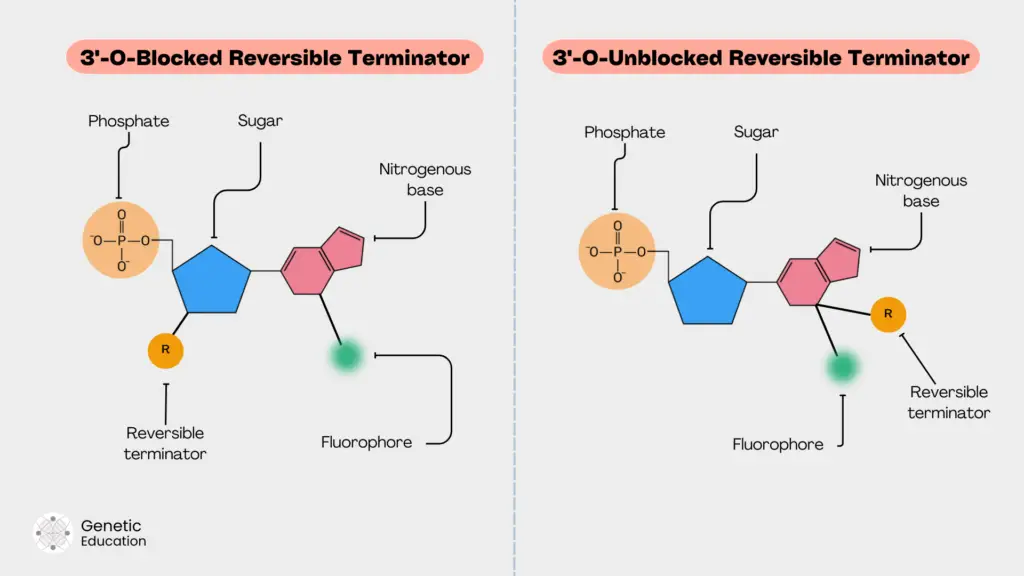
3’-O blocked reversible terminators:
This approach is simple and effective. The blocker (reversible terminator) is directly attached to the oxygen atom of the 3’ OH end of the sugar. This will block the 3’ OH site for polymerase to perform primer extension. Due to this, the blocker can effectively perform the termination.
Examples of 3’ blocked reversible terminators are 3’- ONH2, 3’-O- allyl and 3’-O-azidomethyl.
The present reversible terminator is extensively used by Illumina platforms and provides 100% efficiency for 3’ blocking.
3’-O unblocked reversible terminators:
In this type of reversible terminator, the blocker is directly attached to the base along with the fluorochrome. Due to this, the polymerase can effectively recognize the base for incorporation as it lacks any additional nucleotide structural changes.
An example of the 3’ unblocked reversible terminator is the ‘virtual terminator’ by Helico BioScience Corporation. Another blocker in this category is the ‘lightning terminator.’
Wrapping up:
The reversible terminator is the most excellent example of how we can optimize the product for efficient output. I guess, Illumina is succeeding in this industry because of adapting this technology.
Nonetheless, a reversible terminator has its own challenges to use. I hope you like this information. This is another article of our NGS series. Do subscribe to our blog and share this article.
Sources:
Chen F, Dong M, Ge M, Zhu L, Ren L, Liu G, Mu R. The history and advances of reversible terminators used in new generations of sequencing technology. Genomics Proteomics Bioinformatics. 2013 Feb;11(1):34-40. doi: 10.1016/j.gpb.2013.01.003. Epub 2013 Jan 23. PMID: 23414612; PMCID: PMC4357665.
Bentley, D., Balasubramanian, S., Swerdlow, H. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008). https://doi.org/10.1038/nature07517.


