“This eye-opening article explains which skills you need to learn after 2025 instead of conventional wet lab genetic skills and why they won’t remain relevant.”
ThermoFisher introduced the Genexus NGS system, a fully automated, two-touch, high-throughput, and fast system that eliminates the need for DNA extraction, library preparation, sample preparation, and even analysis.
This is a red flag for anyone planning their career in life sciences. Wet lab work won’t remain relevant post-2025. Read this article to get insight into new technologies, automations, and what skills you need to acquire.
Stay tuned.
Key Topics:
What are wet lab skills?
Wet lab skills are hands-on expertise or practical knowledge to handle chemicals, instruments, reagents and utilities in a biological lab. For instance, preparing a buffer, reaction or chemical, or simply pipetting.
Try understanding the concept by taking an example.
Let say, you want to understand the role of the COMT gene in mental retardation, just assume!
This is your research roadmap.
- Collect the patient samples.
- Prepare DNA extraction chemicals.
- Perform DNA extraction.
- Perform qualitative and quantitative analysis using Nanodrop Lite.
- Design PCR primers and order them.
- Set up a PCR reaction and run the PCR.
- Prepare the gel and run the amplicons.
- Analyse the gel using software or a Gel Doc system.
- Send the amplicon for sequencing. Or prepare the sequencing reaction and run it.
- Collect the data and analyze it.
- Perform BLAST and other analyses to collect meaningful insights.
Now, in this experiment, let’s see which events are considered in the wet lab and dry lab work.
| Wet lab work | Dry lab work |
| Collect the patient samples. | Perform qualitative and quantitative analysis using Nanodrop Lite. |
| Prepare DNA extraction chemicals. | Design PCR primers and order them. |
| Perform DNA extraction. | Analyse the gel using software or a Gel Doc system. |
| Set up a PCR reaction and run the PCR. | Collect the data and analyze it. |
| Prepare the gel and run the amplicons. | Perform BLAST and other analyses to collect meaningful insights. |
| Send the amplicon for sequencing. Or prepare the sequencing reaction and run it. |
Put simply, wet lab work involves physical experiments using chemicals, pipettes, and samples—skills that require precision and hands-on training. In contrast, dry lab work is computational, relying on software and data analysis tools to interpret results.
The table below shows the basic wet lab and molecular biology skills.
| Wet Lab Work Basics | Molecular Biology Skills |
| Pipetting | PCR (Polymerase Chain Reaction) |
| Solution preparation | Gel electrophoresis |
| Weighing chemicals | DNA/RNA extraction |
| Sterile technique | qPCR (Quantitative PCR) |
| Autoclaving | Reverse transcription |
| Buffer preparation | Cloning |
| Centrifugation | Plasmid isolation |
| pH measurement | CRISPR-Cas9 editing |
| Using lab balances | Ligation |
| Labeling and recordkeeping | Transformation |
| Operating incubators | Western blotting |
| Using water baths | Northern blotting |
| Cleaning glassware | Southern blotting |
| Waste disposal protocols | DNA sequencing (Sanger/NGS) |
| Micropipette calibration | Electroporation |
Now! You are wondering, skills like pipetting, solution and reagent preparation, extraction, reaction preparation, etc, are important and need to be learned to work in a genetic lab. Whereas, the analysis work has been reserved to the ‘analysis guy- the bioinformatician.’
You are completely right! I’m not denying.
That’s exactly what your college and professors taught you, and you should have to learn these skills first.
State-of-the-art research and diagnostic laboratories have excluded the manual preparation approach. Automation has already replaced most of the wet lab skills in a couple of years. And mark my words, soon, these skills will be completely replaced by AI and automation.
Why Your Wet Lab Skills May No Longer Be Relevant After 2025
In the era of high-throughput and precision analysis, speed, accuracy and data are the game changers. Imagine how tedious it is to extract DNA from 96 samples within a few hours with consistent accuracy?
KingFisher Flex, QIAGEN QIAcube or MagnaPure can extract DNA from 96 samples in a single run within ~2 hours and with great accuracy. Those belong to the automated DNA extraction unit category.
This is only one example; automation and kits are replacing nearly all the wet lab operations in the genetic lab. Let’s get insight into how our entire working SOP will be replaced by automation, completely.
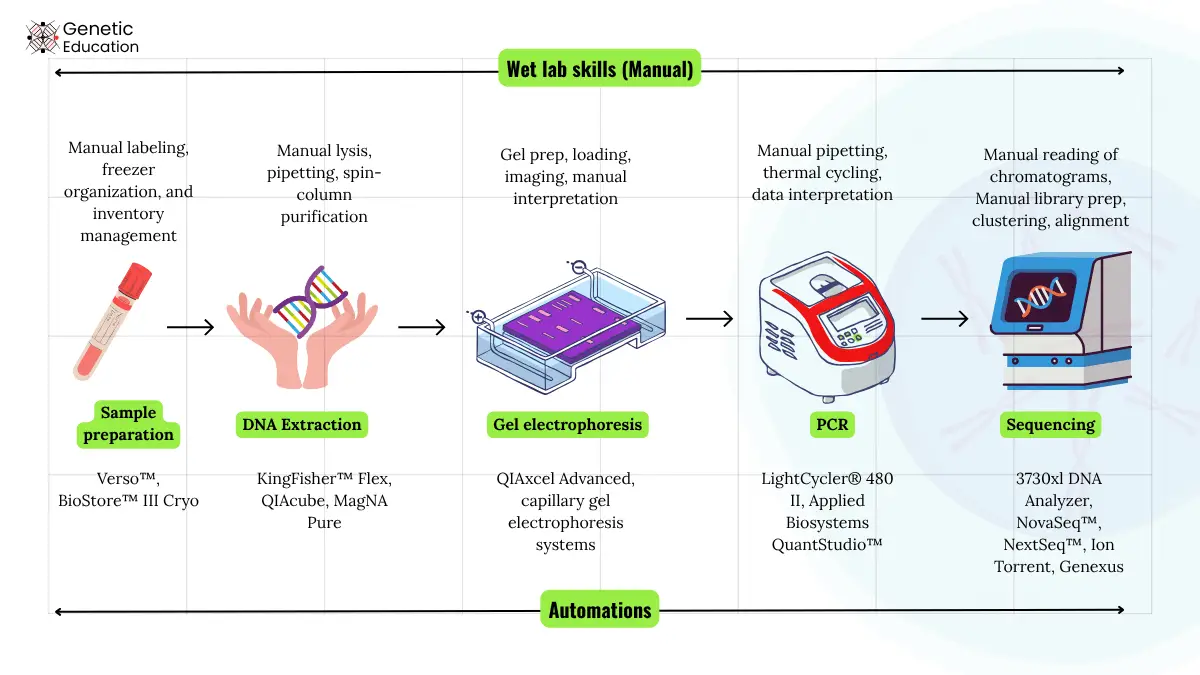
Sample collection:
Sample collection, labeling, storing and managing are manual operations typically performed by lab personnel. Hamilton Verso is an automated sample storage and management system for all biological samples.
It is a fully automated storage, retrieval, shortening, labeling, and tracking system. It can even maintain the cold chain and sterility. It is now widely used in genetic, stem cell, genomic, forensic and other biological labs.
So, no person is needed for manual sample assessment.
Extraction and preparation:
Nucleic acid extraction is a crucial step in the molecular genetics field. It required reagent preparation, extraction and analysis using a gel. And all these steps are manual. You need to
- Prepare solutions like lysis buffer, TE buffer etc.
- Perform extensive pipetting.
- Mixing and centrifugation.
- Gel electrophoresis.
All these steps are manual and have already been mentioned above.
Now, these steps are replaced by the automated DNA extraction units. We just need to insert the sample, that’s it. All steps are performed using a robotic arm and an automated closed system. And will give us pure DNA.
Automated systems are consistent, high-throughput, accurate and widely accepted, and thus gaining popularity.
No additional reagents or kits needed.
In addition, for routine analysis, there is no need to invest time in preparing solutions and buffers; all types of buffers are now available in a ready-to-use form.
Gel Electrophoresis:
Electrophoresis is yet another crucial scheme in a genetic lab as it can separate fragments based on their charge and mass. Capillary gel electrophoresis and eGel have already replaced the conventional gel electrophoresis scenario.
PCR and qPCR:
PCR enables amplification and quantification. It automated the amplification process, backed by the automated template quantification in the quantitative PCR. The availability of ready-to-use amplification kits, although, doesn’t outrank the need for sample preparation, but reduces the reaction preparation time and process.
Fully automated PCR and qPCR analysis software further doesn’t need manual investigations.
Sequencing and NGS:
Sample and library preparation are crucial steps in sequencing and NGS. It allows effective sequencing of DNA fragments. As discussed, the sample preparation and DNA extraction process are already automated.
ThermoFisher’s Ion Chef system is a fully automated template and library preparation, and chip loading system that prepares the chip for NGS analysis. In addition, an automated Sanger sequencing machine doesn’t even require manual chain termination and analysis.
The two-touch approach with KingFisher Flex → Ion Chef → Genexus system completely revolutionized genetic testing by full-scale automation and eliminating the need for manual preparations or wet lab work.
In addition, the availability of third-party genetic analysis software, kits, and digital and automated micropipettes also reduces the manual sample preparation burden. Check out the table below to know which skills are now replaced by automation.
| Wet Lab Step | Automation/Tool | Company/Example | Wet Lab Skills Replaced |
| Sample Collection & Storage | Verso™, BioStore™ III Cryo | Hamilton, Brooks Life Sci | Manual labeling, freezer organization, and inventory management |
| Tube Handling & Sorting | Mohawk Tube Picker, IntelliXcap | Ziath, Azenta | Manual tube picking, labeling, capping/decapping |
| DNA Extraction | KingFisher™ Flex, QIAcube, MagNA Pure | Thermo, Qiagen, Roche | Manual lysis, pipetting, spin-column purification |
| RNA/DNA Quantification | NanoDrop OneC, Qubit 4 Fluorometer | Thermo Fisher | Manual UV measurement, pipetting, dilution calculations |
| qPCR Setup & Analysis | LightCycler® 480 II, Applied Biosystems QuantStudio™ | Roche, Thermo Fisher | Manual pipetting, thermal cycling, data interpretation |
| Buffer and solutions | Available in ready to use form | – | Manual buffer and solution preparation |
| Gel Electrophoresis | QIAxcel Advanced, LabChip GX | Qiagen, PerkinElmer | Gel prep, loading, imaging, manual interpretation |
| DNA Sequencing (Sanger) | 3730xl DNA Analyzer | Applied Biosystems | Gel prep, manual reading of chromatograms |
| Next-Gen Sequencing | NovaSeq™, NextSeq™, Ion Torrent, Genexus | Illumina, Thermo Fisher | Manual library prep, clustering, alignment |
| Data Analysis | BaseSpace, Geneious, Galaxy | Illumina, Biomatters | Manual alignment, BLAST searches, QC reporting |
In conclusion, your wet lab skills are now the least important in the upcoming time. Worried? Now you may question, what to do instead?
What do you do?
‘We don’t need to learn these skills?’ You might have this question.
The answer is that you should learn! These skills are the basic foundation of genetics or life sciences. But don’t rely only on these skills.
Your wet lab training isn’t sufficient, which students mostly look for! Now, instead, focus on the analysis part. Learn bioinformatics, tools and software to analyze the results.
Learn to manage, optimize and improve the assays. Inaccurate results and consistency are still a challenge. Learn to overcome those problems. You need to now focus on optimizing the assay and not on buffer preparation.
For instance, you have 10 different samples for breast cancer NGS analysis. You need to learn skills regarding how to combine all these 10 samples in a single assay and still achieve accurate results. This saves time and cost.
You need to develop a unique sense or expertise to choose a technology for the lab. For instance, NGS is now a mandatory technology, particularly short-read sequencing for oncology and cancer.
Conversely, long-read sequencing, such as PacBio or ONT, is completely useless for routine diagnosis.
Train yourself with diverse technologies, and understand how and when to use them. The game is now on cost, speed and accuracy.
The era of genomics began; those who work on throughput, speed and cost will win the race, ultimately.
Disclaimer:
Keep in mind, techniques like karyotyping or FISH still highly rely on manual wet lab work. If you want to pursue your career in cytogenetics, you need to sharpen your wet lab skills.
Wrapping up:
In conclusion, your genetics knowledge and practical learning aren’t outdated. You just need to reshape them. Invest your time and money in learning analysis and optimizations. That’s what I can say from my personal experiences.
However, if you don’t wish to transition out of academics, wet lab skills can help you, perhaps. May this article serve as both a guide and a wake-up call as you prepare for a career in genetics.
Do share this article and subscribe to Genetic Education.


I wanted to take a moment to commend you on the outstanding quality of your blog. Your dedication to excellence is evident in every aspect of your writing. Truly impressive!