“Genomic imprinting– A small subset of genes in mammals are inherited only from single parents and are epigenetically regulated during parental germline division.”
Genomic imprinting has been an unknown mechanism for years. However, recently it gained more light. It states– a small pool of genes are mono-allelic and have parent-specific origins. Meaning, from a pair of genes, only a single copy is active.
Imprinting by epigenetic reprogramming occurs during germinal cell division, passes on to offspring by fertilization (maintained), and is erased during the development of a new set of germ cells.
That entire cycle of epigenetic programming and reprogramming is highly regulated in mammals and occurs continuously under the influence of environmental signals. A known epigenetic tag or process is involved in the entire cycle.
Various enzymes of the same family catalyze the cycle of programming, maintaining, and reprogramming. So what’s that epigenetic phenomenon behind genomic imprinting? What are those enzymes and molecular mechanisms using which genomic imprinting occurs?
We have explained the concept of genomic imprinting– in Lyman in our previous article of this series. If you want to learn, you can go and read that article (link is given). Here, we are discussing the molecular mechanism behind genomic imprinting.
Stay tuned.
Molecular Mechanism of Genomic Imprinting
Previous research manifests that the parental genomes are functionally inequivalent which shows that even though both parents contribute the same number of chromosomes and even genes, are not functionally similar.
The differential epigenetic pattern of each parental genome makes it functionally dissimilar. Hence the genome of both parents is required to fertilize and produce a healthy embryo. Imprinting by DNA methylation, a known and proven epigenetic characteristic of the mammalian genome, is the main process behind the functional diversification of some genes.
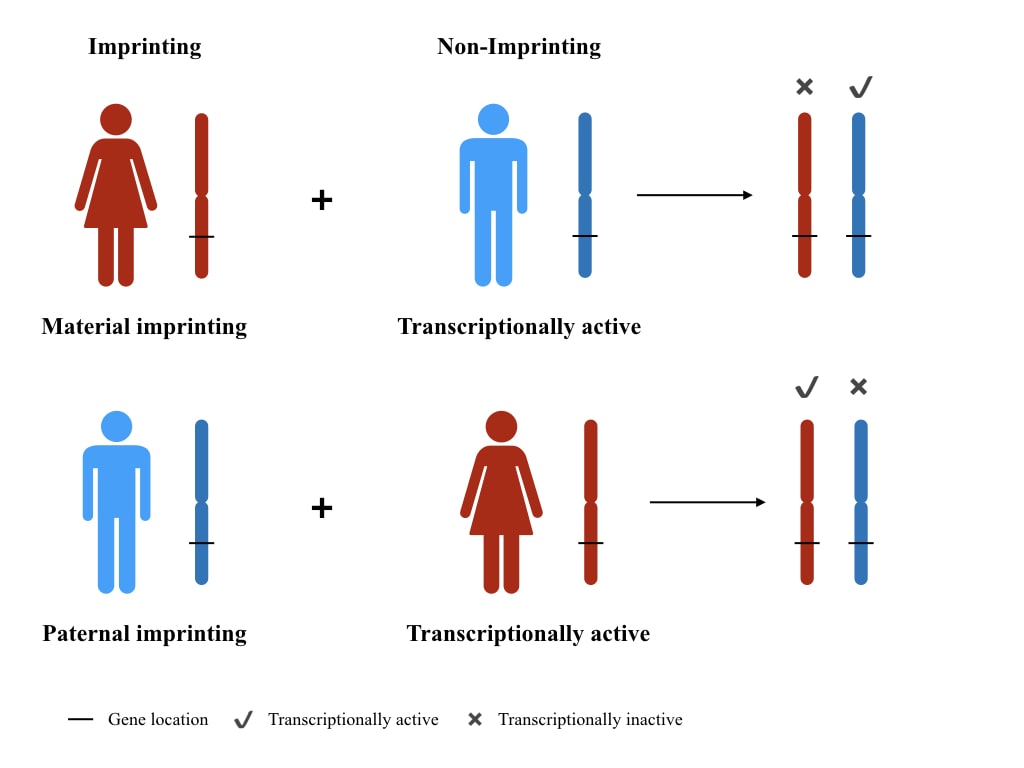
For diploid organisms (like us) both parents equally inherited the majority of genes. However, a small percentage of genes are uniparental– expressed from either the maternal or paternal side. Imprinting makes a gene inactive from a single parent.
For example, if any gene is maternally imprinted, only the parental allele for that gene activates, similarly, if any gene is paternally imprinted, a maternal allele for that gene activates. Broadly, the phenomenon of imprinting is categorized as an epigenetic modification.
‘Epigenetics’ –studies changes in gene expression and not changes in gene structure. Demonstrating, not only the structure of a gene, but the differential expression of that particular gene also contributes to producing diverse phenotypes.
Methylation, acetylation, and histone modifications like histone phosphorylation, ubiquitination, methylation, etc are popular epigenetic mechanisms, well-explained in literature. Those are often known as epigenetic tags too.
DNA methylation is one of the known and common epigenetic tags for the occurrence of genomic imprinting. Enzyme methyltransferases (DNMT) catalyze various steps of methylation and demethylation during the establishment, maintenance and removal of imprinting.
DNA methylation atypically occurs in the CpG-rich region of our genome, where most of the imprinted genes are clustered. The enzyme- methyltransferase adds the methyl group to the C5 residue of the cytosine nitrogenous base and eventually makes it transcriptionally inactive.
What happens here?
When any cytosine base methylates from the cluster of the imprint-Susceptible regions, it disallows transcriptional factors to recognize the gene and perform gene expression. The process is de novo DNA methylation, meaning, fresh/new DNA methylation occurs.
This particularly occurs during either germ or egg development and by the act of de novo DNA methylation, a new and differential epigenetic pattern– genomic imprint generates for each, either sperm or egg.
Studies on mice models demonstrate that DNMT3A and DNMT3B are two methyl transferases that participate in the de novo methylation of DNA during gametogenesis. DNMT3L is yet another protein that supports the mechanism, particularly in maternal imprinting. However, the DNMT3A/DNMT3L complex is essentially required for the imprinting on the maternal side– in oocytes, more studies manifest.
The same complex of proteins is also essentially required for imprinting in male gametes. Albeit, how this complex recognizes whether it’s egg or sperm cell imprinting is still a question for scientists. No clear evidence is reported.
By the act of fertilization of germ cells, a new embryo is formed having differential ‘imprint’ which is notedly, remains ineffective from the global epigenetic reprogramming that occurs during the pre-implantation stage.
This particular step is known as the ‘establishment’. Now the second stage of this process is to maintain the particular imprint throughout the life of an organism which is the ‘imprint maintenance’ stage.
DNMT1 and its isoforms have a pivotal role in maintaining the imprint, not only during the pre-implantation stage but the entire life and in all somatic cells division. The reasons why DNA methyltransferase 1 (DNMT1) is crucial for this process are because it associates with the replication fork and is a strong substrate for hemimethylated DNA too.
DNMT1 ensures, if methylation of DNA once occurs, will remain forever in somatic cells, and at least until the second round of imprinting. To support this evidence, a study on knockout mice for the Dnmt1 gene demonstrates a lack of imprinting.
The maintenance process governed by the DNMT1 has been reported in most imprinted genes in mice and humans too.
Other secondary proteins like ZFP537, methyl-CpG-binding protein and PGC3 (Stella protein) prevent maternal and paternal imprints from being reprogrammed during preimplantation epigenetic reprogramming.
The maintenance of imprints in somatic cells (embryogenesis) is also regulated by DNMT1. Studies proclaim that other epigenetic modifications like histone modifications also get involved in this process.
In fact, the DNMT1 govern methylation is less likely to be important for maintenance in somatic cells. The collaborative effort of such mechanisms tightens the chromatin by disallowing transcriptional factors and transcription to occur.
Now at the very end of the process, to produce a new imprinting pattern, the newly formed germ cells must undergo de-imprinting or demethylation. Demethylation is catalyzed by demethylase enzymes.
During primordial germ cell division by which new sperm or egg cells form, TET family proteins govern the process of demethylation. Various TETs perform various functions, for example, detachment of DNMT1 and the process of DNA demethylation.
This stage is known as the ‘erasing of imprinting’. The entire process of erasing the imprint is a collective effort by DNA demethylation and DNA repair mechanism.
Note
DNA methylation isn’t the only and necessary epigenetic process that produces genomic imprinting. In the absence of methyltransferase enzymes, imprinting also occurs (Dalgaard and Klar, 1999 and Wolffe and Matzke, 1999).
The clear objective of imprinting to occur in germ cells is to disallow a gene for transcription; protein may not form, consequently. DNA methylation– a popular way for genomic imprinting can make an imprinted gene transcriptionally inactive by two mechanisms.
First, The added methyl group directly interferes with transcriptional factors by blocking the site for various factors.
Second, the methylation recruits other proteins like methyl-binding domain proteins (MBD1 and MBD3) and methyl CpG- binding protein 2 (MeCP2) for transcriptional repression.
Wrapping up:
The molecular mechanism of genomic imprinting majorly involves the process of methylation or epigenetic silencing, methyltransferase enzymes and other supportive proteins. The imprinting completes in three distinct steps.
- First, the establishment of imprints by DNA methylation (DNMT3A and DNMT3B) during germ cell development.
- Second, maintenance of imprinting by DNA methyltransferase 1 (DNMT1) during preimplantation reprogramming and during somatic division.
- Third, erasing or removing the previous imprint to reprogram the new genomic imprint into the germ cells (TET).
I hope you like this article. If you want to understand the concept very well, you can read our previous article.
Resources:
Li, Y., Sasaki, H. Genomic imprinting in mammals: its life cycle, molecular mechanisms and reprogramming. Cell Res 21, 466–473 (2011). https://doi.org/10.1038/cr.2011.15.
Pfeifer, K. “Mechanisms of genomic imprinting.” American journal of human genetics vol. 67,4 (2000): 777-87.
Dalgaard, J Z, and A J Klar. “Orientation of DNA replication establishes mating-type switching pattern in S. pombe.” Nature vol. 400,6740 (1999): 181-4.
Wolffe, A P, and M A Matzke. “Epigenetics: regulation through repression.” Science (New York, N.Y.) vol. 286,5439 (1999): 481-6.