“Cpf1 previously known as CAS12a is a type of effector nuclease, used in the CRISPR system that uses only the (guided) crRNA for catalytic activity.”
CRISPR is an integral part of the prokaryote defense system that uses nuclease as Cas protein to destroy phage DNA or other foreign invaders. CRISPR-Cas is a common name for the system and gained popularity for its excellent nuclease efficiency.
Various CRISPR systems have been reported in different bacteria and archaea and are broadly categorized into types I and II; depending upon the Cas nuclease the system has.
Every modified Cas has a significantly different function, for example, the dCas9 has a role in gene activation and repression. Cas12a or Cpf1 also has crucial functionality, especially in plant genome editing.
Here in the present article, I will explain about Cpf1-CRISPR system, its structure and its importance in gene therapy. We are discussing every minute detail regarding CRISPR (No matter how small the search volume is!) to strengthen your CRISPR knowledge.
Read more articles on this topic: CRISPR-CAS9.
Stay tuned,
Key Topics:
What is CRISPR-Cpf1?
Cpf1 stands for CRISPR from Prevotella and Francisella 1, belongs to subtype V from the bacterial CRISPR system. Previously it was known as Cas12a, however, it works similarly to other Cas proteins.
The Cpf1-mediated CRISPR system is widely used in plant genome editing because of some tremendous advantages over Cas9 (that we will discuss later). Let us first understand the structure of Cpf1.
| Cpf1 | CRISPR from Prevotella and Francisella 1 |
| Size | 1200 to 1500 amino acids long |
| PAM sequence | 5’- TTN -3’ |
| Alternative name | Cas12a |
| tracrRNA | Not required |
| Nuclease activity | dsDNA cut at two different locations |
Structure:
Nuclease usually has two domains; DNA binding domain/ DNA recognition domain and nuclease domain. The binding domain settles on DNA and binds to the target location while the nuclease domain cleaves the DNA which is usually a dsDNA cut.
Cpf1 has two lobes or domains; REC and NUC short of helical or RNA recognition lobe and nuclease lobe, respectively. REC consists of two more subdomains- REC1 and REC2 with 13 alpha helices and 19 alpha & two beta chains, respectively.
Contrary, the NUC comprises RuvC, WED, Pi and Nuc domains. Every domain has a different function.
| Type of Cpf1 | Length (aa) | PAM sequence |
| LbCpf1 | 1228 | 5’-TTTV -3’ |
| AfCpf1 | 1307 | 5’-TTTV -3’ |
| FnCpf1 | – | 5’-TTN -3’ |
The structural analysis of Cpf1 and Cas9 explained that the common and conserved RuvC domain has a nuclease function derived from autonomous transposons like the IS605.
Cpf1 seed region:
The Cpf1 seed region is a 5 to 6 nt long region at the 5’ end near the spacer region which is required when crRNA-target DNA hybridization occurs. Put simply, the seed region locates the complex on the target by complementary base pairing.
PAM for Cpf1:
The PAM sequence region has important significance in in vivo as well as in vitro gene editing, however, depending upon the system, the structure and nucleotide composition of PAM vary.
You can learn more by reading this article: PAM (Protospacer Adjacent Motif) sequence.
The PAM for Cpf1 is a T-rich region with a usual sequence structure 5’- TTN -3’. Importantly, the PAM recognition is completed using the conserved lysines of WED and PI domains of the protein.
Various Cpf1 proteins and their PAM sequence is enlisted in the table above.
Important information:
The Cpf1 also known as the Cas12a has both RNase and DNase activity which allows it to cut the CRISPR locus and the target dsDNA.
Function:
In a natural system, Cpf1 binds to the single guided RNA and at a target location upstream to the PAM sequence, make a double-stranded 5 bp staggered cut and cleave off the DNA.
Here the REC lobe finds and binds to the crRNA-DNA heteroduplex junction while various subdomains of NUC facilitate effective cutting. The PI domain of the NUC lobe interacts with the PAM sequence (Protospacer adjacent motif) while the NUC performs a catalytic reaction. Notedly the function of other NUC domains is unknown.
Cpf1 vs Cas9
Cas9 is one of the most widely used nucleases in the CRISPR technology, however, the Cpf1 has several groundbreaking advantages over the Cas9. Check out several common differences between each.
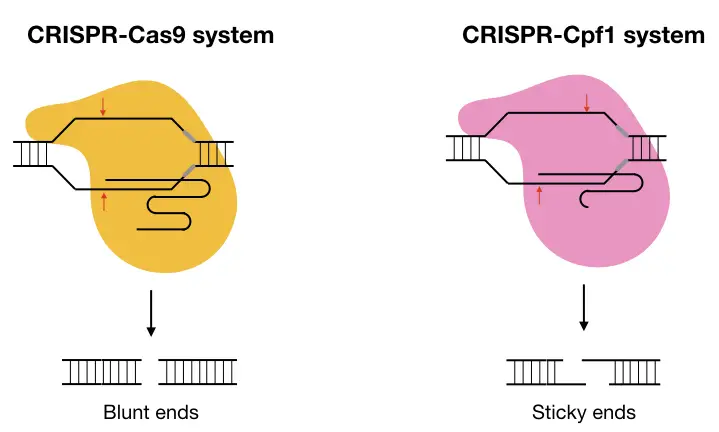
The catalytic activity of the Cas9 highly relies on the sgRNA which is the composition of two individual RNA- crRNA and tracrRNA, so the Cas9 needs two RNAs while the Cpf1 requires a single RNA as crRNA.
Moreover, Cas9-dependent crRNA used in the gene therapy is comparatively longer in size than the crRNA required in Cpf1-mediated therapy.
Studies also revealed that the cuts generated by the Cas9 and cpf1 produce blunt and staggered ends, respectively. The Cas9 cleaves at the same location on two different target strands while the cpf1 cleaves at two different locations and produces 5bp long staggered ends.
One other difference explains that the PAM region for Cas9 is usually G-rich while the PAM region for Cpf1 is usually a T-rich sequence. In addition to this, the cutting sites for Cas9 and cpf1 are proximal and distal to the PAM sequence, respectively.
One article of Wikipedia also explains that the use of CRISPR-Cas9 is subjected to IPR while the Cas12a or Cpf1-mediated gene therapy doesn’t have the same issue.
The capf1 has more strong multiplexing outcomes than the cas9 in which a single crRNA can be used to edit multiple genomic regions.
Structurally, Cas9 has two nuclease domains viz RuvC and HNH while cpf1 has a single RuvC like endonuclease domain.
Applications of CRISPR-Cpf1 in genome editing:
Much like Cas9, the Cpf1-mediated CRISPR can also be used as a gene activator and repressor. Designing the dCpf1 or dCas12a without the catalytic activity can be used directly as a repressor or utilized as a vehicle to transfer gene activator for triggering transcription.
Tang et al., 2017 performed repression of miR159b and reduced expressions many folds in rice and Arabidopsis.
Yin et al., 2017 also reported a higher deleterious mutagenesis effect of LbCpf1 like CRISPR-cpf1 system in plants of up to 63bp deletion.
Other comprehensive studies also reveal that transformation of Cpf1 using particle bombardment, PEG and classic plant gene delivery vector- Agrobacterium gives excellent results as the size of the coding gene segment of Cpf1 is very less.
Studies on tobacco and soybean also suggest that Cpf1 protein can directly be used in DNA-free editing. Here a purified cpf1 protein along with the guided RNA can be directly transferred at the target (Kim H et al., 2017).
More applications are enlisted here:
| Plant species | Target gene | Transformation technique |
| Rice | EPFL9 | Agrobacterium |
| Arabidopsis | OsDEp1 | Floral dip and protoplast transformation |
| Soybean, tobacco | FAD2-1A and FAD2-1B | PEG-mediated protoplast transformation |
| Rice | DL, ALS, NCED1 | Agrobacterium |
Advantages of CRISPR-cpf1:
The use of Cpf1-mediated CRISPR technology has more advantages in plant genome editing than the conventional Cas9.
- It required shortened sgRNA.
- It doesn’t need the tracrRNA.
- It can be used for DNA-free editing.
- It can be used as a repressor & activator and can produce larger deleterious mutations.
- The shorter coding sequence increases the efficiency of the delivery vector many folds.
- One of the biggest advantages of the present system is the multiplexing, Cpf1 allows researchers to edit multiple loci using a single system.
Multiplex editing using CRISPR-Cpf1:
Cpf1 can do editing at many different loci in a genome, studies on plant genome explain using two approaches; expressing various sgRNA using single promoter and expression various sgRNA using multiple promoters.
Scientists use the second approach in which separate single sgRNA along with individual promoters can be transferred and expressed using the same or different vectors.
Limitations of CRISPR-Cpf1:
Every technology comes with shortcomings and so does the cpf1 too. One important property of cpf1- a shorter crRNA which is the advantage is the limitation as well.
Shorter crRNA has drawbacks like it can form secondary structure easily, decrease efficiency and effectiveness.
Furthermore, to use it in plant genome editing, it needs a standard temperature setup to work efficiently.
Wrapping up:
Undoubtedly, the cpf1-mediated CRISPR system has immense applications in plants however, it’s yet not ready to use on mammals. Studies also explain that it can be used for multiplex editing using shorter crRNA.
The technique is temperature sensitive and so various systems of cpf1 need separate temperature setups. Conclusively, though cpf1 is more advantageous, it’s less effective than cas9.
Sources:
- Safari F, Zare K, Negahdaripour M, Barekati-Mowahed M, Ghasemi Y. CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci. 2019;9:36. Published 2019 May 9. doi:10.1186/s13578-019-0298-7.
- Alok A, Sandhya D, Jogam P, Rodrigues V, Bhati KK, Sharma H and Kumar J (2020) The Rise of the CRISPR/Cpf1 System for Efficient Genome Editing in Plants. Front. Plant Sci. 11:264. doi: 10.3389/fpls.2020.00264.
- Kim, H., Kim, S. T., Ryu, J., Kang, B. C., Kim, J. S., and Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8:14406. doi: 10.1038/ncomms14406.
- Kim, H. K., Song, M., Lee, J., Menon, A. V., Jung, S., Kang, Y. M., et al. (2017). In vivo high-throughput profiling of CRISPR–Cpf1 activity. Nat. Methods 14, 153–159. doi: 10.1038/nmeth.4104.
- Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., et al. (2017). A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3:17018.
- Yin, X., Biswal, A. K., Dionora, J., Perdigon, K. M., Balahadia, C. P., Mazumdar, S., et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757. doi: 10.1007/s00299-017-2118-z.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.