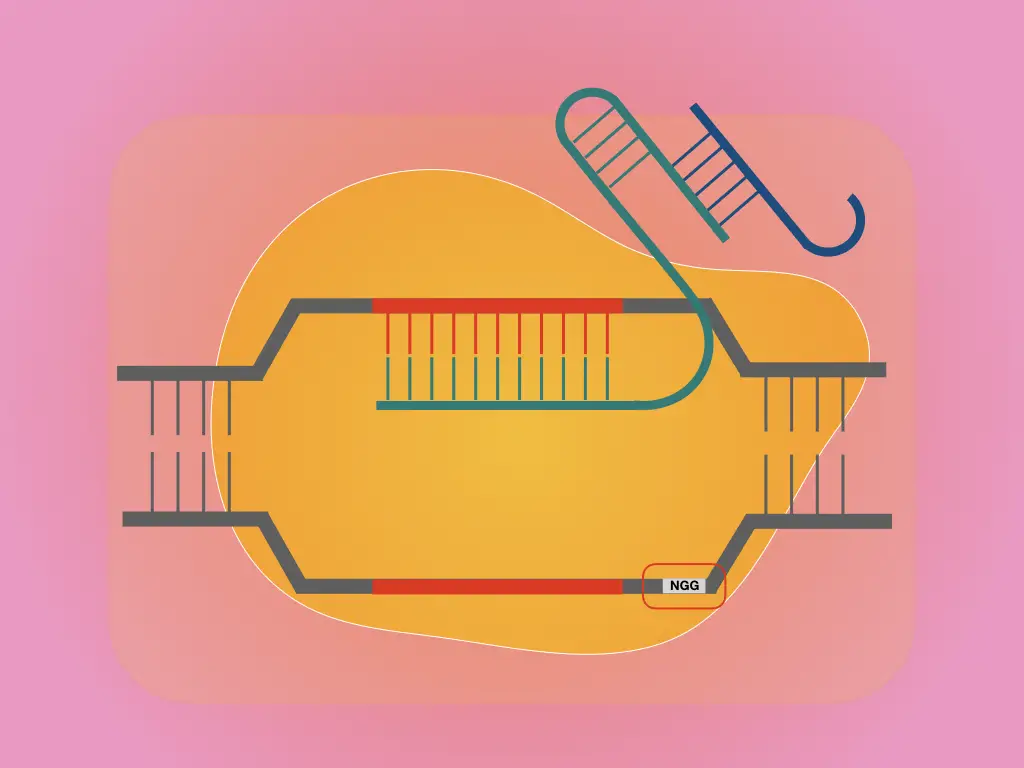
“PAM stands for Protospacer Adjacent Motif, which functions as two-factor authentication for Cas to cleave the foreign invading DNA only.”
The CRISPR system has been an integral part of bacterial immunity, perhaps working for thousands of years. It highly relies on the CRISPR locus and Cas protein, largely, however, the system needs other miniature elements to fine-tune functions. For example, the leader sequence to transcribe the Cas gene cluster and PAM sequences for accurate target localization.
Roughly, the series of ‘Cases’ completes the whole process for integration to interference. A type of Cas cleaves off phage DNA and integrates it into the host CRISPR followed by crRNA transcription. Another class of Cas binds with the mature crRNA, uses it as an identifier and cleaves off the target DNA.
If you understand CRISPR and are a die heart fan! You may wonder… wait, something is not right!
The spacer sequence present in the CRISPR array has been derived from the invading bacteriophage or plasmid and used as a memory unit. Meaning, it’s exactly identical to some sequence of a phage.
So if the Cas can cut the foreign invading DNA using the memory of a spacer, it can cut their own DNA too (more precisely at where the spacer is located). Your doubt is valid even though.
Hypothetically, it’s a correct situation much like the restriction digestion mechanism.
But there is a catch: they can prevent these types of self-destruction activities using the ‘second-factor authentication system’ which accurately investigates the intruder DNA and validate cutting activity.
PAM sequence has been well-known as a second-factor authentication for identifying the specific invading DNA, discriminating self vs non-self DNA and cleaving off the target DNA only.
To better comprehensively understand CRISPR technology, I am writing this article on PAM, hoping to improve your overall CRISPR knowledge. But before that, if you are a CRISPR fan! Check out the entire category:
Key Topics:
What is the PAM sequence?
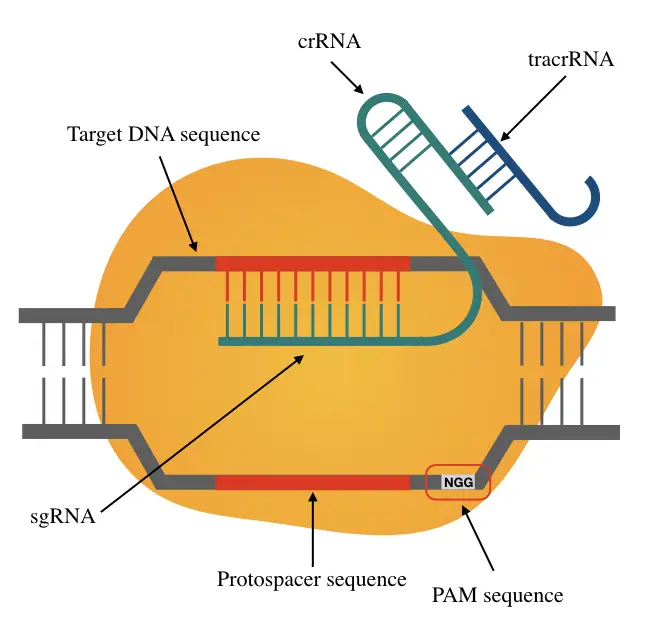
PAM short for Protospacer Adjacent Motif is up to 6nt short sequences recognizable to Cas for target-specific nucleophilic activity. Precisely, It’s 2 to 6nt long and located downstream to the protospacer region on the intruding DNA.
Note that the PAM is located 3 to 4nt downstream to the nuclease cleavage site.
In a natural system, Cas1 and Cas2 recognize PAM during the spacer acquisition; it can’t incorporate spacer in their CRISPR until it finds a specific PAM sequence to target (Richter C and Dy R et al., 2014).
Jinek M & Chylinski K et al., (2021) and Sashital D & Weidenhelf B (2021) explained that the functional PAM domain can be further separated into SAM- spacer acquisition motifs and TIM- target interference motifs.
SAM is involved in the spacer integration process while TIM is involved in the recognition domain during the interference process.
Read more: Applications of CRISPR-CAS9 in Medical Science, Diagnostics, Research, Plant Biology and Agriculture.
Properties of PAM sequence:
- The PAM is a single, a few base pairs long sequence present on the protospacer region of the invading DNA.
- PAM is present on the target phage or plasmid.
- PAM isn’t a part of the bacterial immune system.
- The spacer present in the bacterial CRISPR does not have PAM.
- The protospacer contains the PAM and is present on the target phage or plasmid.
Examples of PAM Sequence:
| PAM sequence | Organism |
| NGG | S. pyogenes |
| NGRRT | S. aureus |
| NNNNRYAC | C. jejuni |
| NNNNGATT | N meningitidis |
| CCN | S. solfataricus I-A1 |
| TCN | S. solfataricus I-A2 |
| NAAR | S mutan |
| TTN | A acidophilus |
| TTTV | Acidaminococcus sp. And Lachnospiraceae bacterium |
| NNAGAAW | S thermophilus |
| E coli | CC |
| E coli | AWG |
Role of PAM in spacer acquisition:
Recalling the process of CRISPR in bacteria, spacer acquisition has been known for the incorporation of invading DNA into the CRISPR locus.
Previous advancements in the CRISPR field suggest that not only the Cas but also the leader sequence, repeat region and the PAM sequence play a significant role during acquisition (Shah A, Hansen N & Garrett R, 2009 and Shah A & Edder P et al., 2009).
The in-depth analysis explains that PAM provides the recognition site for Cas to cleave protospacer. Loss of mutation studies evident that when a PAM has been not identifiable or lost, the acquisition process abrupts.
In addition, protospacers also exhibit PAM internally within their sequence for other protospacer (for other CRISPR loci). The whole process and role of PAM in integration have yet not been well explained.
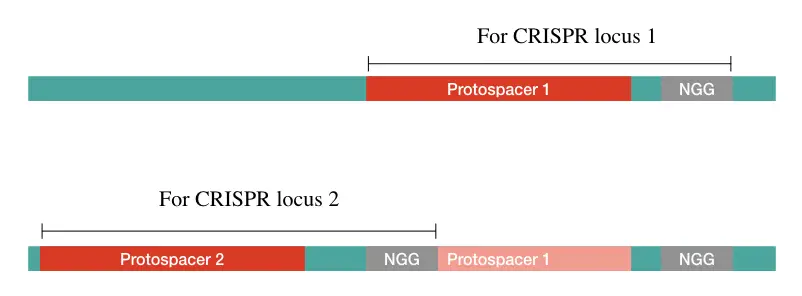
Put simply, PAM works as a marker for the Cas to recognize the protospacer, cleave it and integrate it into the CRISPR.
Read further:
Role of PAM in interference:
In a bacterial defense system, the interference process collectively destroys the foreign DNA by a nucleophilic attack. A Series of works explains that PAM hasn’t a significantly important role in interference.
However, insertional mutagenic studies suggest that in absence of a specific PAM or mutant PAM, the mechanism can’t work and the virus can easily skip off the mechanism.
Contrary studies also explain that except for a few bases, when a mutation occurs in a non-conserved location on PAM, the interference still works (Gracia-Heredia I & Marin-Cuadrado AB, 2012; Lopez-Sanchez MJ, Sauvage E).
The present work has been supported by Almendras et al., (2012) stating that the presence of the Cytosine nucleotide at positive 2 and 3 in the PAM greatly increases interference activity.
Put simply, the role of PAM exclusively can’t be explained or denied in the process of interference. More research is required to support either statement.
Collectively, it is said that PAM has an important role in the CRISPR system and was established intentionally. It’s an important marker-like region for Cas to perform endonuclease action- in acquisition or interference.
So one can ask if it has any role in CRISPR-mediated gene editing? Or can create problems during gene editing? Let’s find out.
Important point:
Keep in mind that PAM helps in recognition but is not included in the cleavage process.
Related article: How does the CRISPR CAS9 System Distinguish Self vs Nonself DNA?- 7 Proven mechanisms.
Role of PAM in gene editing:
Using the spacer sequence, CAS can work but the presence of the PAM sequence adds an extra layer of precision and accuracy to perform editing. So hypothetically and practically, the need for PAM makes gene editing less tensile.
Yes, it creates a problem. Because a gene of interest (What we are experimenting on) doesn’t necessarily have a PAM sequence. If so, it would be coincidental. However, unlike the bacterial own system, the presence of PAM adds another level of precision.
So it overall makes the gene therapy even more accurate than PAMless editing. But the question is, it’s impossible, what sequence we choose to acquire the PAM. As we talked about in the above section, various Cas have different PAM sequences. The list has been given above.
It depends on the Cas derived from the bacteria. And may vary. So during the gene therapy, first researchers have to locate or identify the PAM-like sequence, analyze it and correlate it with available Cas options.
That’s the only solution at least for now. And it makes the technology accurate but limited. Scientists prepared synthetic Cas protein and modified naturally occurring protein as per the requirement.
The moral of the story is that using the PAM is a wise decision to achieve precision in edition, greatly. Interestingly, using PAM can be a wiser decision as well, in a few cases.
Reverse engineering suggests that incorporating the PAM in the DNA sequence of gRNA is useful in cell differentiation and cell barcoding process.
Here, a guided RNA manufactured from the PAM containing DNA destroys their own DNA thereby useful in cell lineage study often known as homing sgRNA. Homing gRNA is an interesting topic, we will discuss it in some other article.
Walton et al., 2020 recently explained nearly a PAMless system that targets a whole-genome rather than a single locus. Their findings state that the combined effect of SpRY and the SpG like system eliminates the need for PAM.
And therefore can target many locations, genes or sequences using a single CAS without requiring the PAM. Notwithstanding, the efficiency and accuracy of these types of systems aren’t yet accessed.
Note
PAM isn’t relevant in RNA cleavage. It’s only used to cleave DNA by the Cas.
Wrapping up:
“PAMless is useless”.
A CRISPR Cas9 system must need a PAM to perform its function effectively. So the traditional thought of giving more importance to Cas is not completely true. The system needs a Protospacer motif to even start editing both in vivo and in vitro.
Currently, no specific option eliminating the need for PAM is fully explained. However, scientists are working hard to develop one.
Hope so in the future, we would develop an advanced system that uses a single Cas to eliminate the need for PAM and can efficiently work for the whole genome.
FAQs:
Sources:
Almendros C, Guzmán NM, Díez-Villaseñor C, García-Martínez J, Mojica FJ. Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli. PLoS One. 2012;7(11):e50797. doi: 10.1371/journal.pone.0050797. Epub 2012 Nov 26. PMID: 23189210; PMCID: PMC3506596.
Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 2012 Sep;85(6):1057-71. doi: 10.1111/j.1365-2958.2012.08172.x. Epub 2012 Jul 27. PMID: 22834929.
Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009 Feb;37(Pt 1):23-8. doi: 10.1042/BST0370023. PMID: 19143596.
Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009 Apr;72(1):259-72. doi: 10.1111/j.1365-2958.2009.06641.x. Epub 2009 Feb 23. PMID: 19239620.
Garcia-Heredia I, Martin-Cuadrado AB, Mojica FJ, et al. Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS One. 2012;7(3):e33802. doi:10.1371/journal.pone.0033802.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.