CRISPRi, short for CRISPR interference, is designed to repress gene expression using the customized sgRNA and dCAS.
CRISPR- CAS9 is simply amazing!
Why?
Because it can edit a gene at a specific location, not only that it has other outstanding applications as well, for example, regulating gene expression.
That’s why I love CRISPR, if you are new to this, I strongly recommend you to read this article before going ahead with the present topic:
Put simply, by incorporating the target sequence into the CRISPR, designing the customized CAS protein and target-specific guided RNA, a sequence or gene can be effectively edited.
If you have read the above articles, you now understand that there are tons of variations of the native system exist, and are used for various purposes.
In recent times, scientists are using various CRISPR-CAS assays for regulating gene expression, which literally gives outstanding results. CRISPRi is one such technique to repress gene expression.
The present blog majorly focuses on CRISPRi or CRISPR interference, what it is and how it is constructed. I will also discuss applications of the present technique and will share some other excellent information as well.
We are exploring CRISPR and related technology. I have written 5 articles to date that you can read by revisiting this category:
I hope the present article will add more value to your CRISPR knowledge and help you to understand it better.
Do you know?
40% bacterial and 90% archaea consist of the natural CRISPR mechanism as a part of their defense system.
Key Topics:
What is CRISPRi or CRISPR interference?
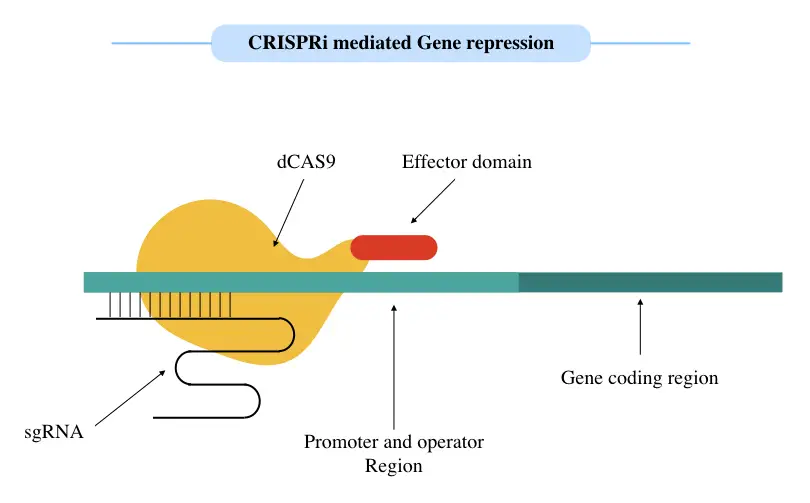
CRISPRi or CRISPR interference is an artificial gene regulation system that uses the dCAS9 protein along with transcriptional factors to repress the expression of a specific gene or set of genes.
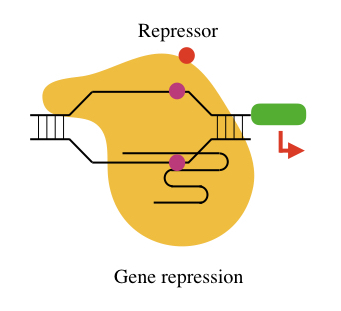
Though we have a well-established RNA interference (RNAi) system to regulate gene expression, there is a major difference between CRISPRi and RNAi. The RNA interference regulates gene expression at the mRNA level while the CRISPR interference technique regulates gene expression at the transcription level.
The CRISPRi technique was first studied at the University of California in San Francisco by Stanley Qi in 2013.
Studies indicated that stronger repression action is observed in both prokaryotes and eukaryotes, notedly, up to 90%, effective gene silencing was reported in the human cells using the CRISPRi technique.
Important terminologie:
sgRNA: sgRNA is a single-stranded guided-RNA or guided RNA that navigates the CAS9 at the target location. It has two parts tracrRNA and crRNA.
CAS9: The CAS9 or CAS is an endonuclease enzyme that cleaves DNA at a target location.
dCA9: dCAS9 is a dead CAS protein that lacks endonuclease nucleophilic activity. It can’t cut DNA.
As we talked, the present technique blocks/abrupt transcriptional activity at the transcription level, meaning, during the process. It indeed blocks either initiation or elongation step of the transcription.
Mechanism of action:
Along with the CRISPR sequence; dCAS- dead CAS and synthetic guided RNA have been utilized for the repression action. Here, the dead CAS though does not have the endonuclease domain but has the DNA binding domain which helps the repressor to locate on a DNA or gene at a specific location.
The 20nt target sequence-specific part of the sgRNA guides the dCAS at a specific location, note that the dCAS binds at the 43 nt CAS binding site on the sgRNA.
The complex delivers the transcriptional repressor or transcriptional effector molecule to the exonic or promoter region of a gene which when located blocks the binding site for other transcriptional activation proteins.
Resultantly, the RNA polymerase can’t recognize, bind and synthesize the strand until it finds transcriptional activators. It ultimately aborts the process.
Among two types of CRISPRi mediated repression (template strand-specific & non-template strand-specific), non-template strand-specific repression gives excellent results when using the dCAS. Studies suggest that 80% to 90% repression efficiency was reported in eukaryotes using the non-template strand-specific dCAS mediated CRISPRi.
The binding of the dCAS to a gene prevents the association of cis-acting and trans-acting transcriptional factors thereby disrupting the process.
Noteworthy, this works effectively at transcription initiation and elongation level.
Another method to suppress gene expression is artificial heterochromatinization. Active euchromatin can be artificially converted into heterochromatin by practicing dCAS fused effector or repressor domain.
An excellent example is the KRAB domain, when fused with the dCAS it allows strong repression of the targeted gene.
It’s important to keep in mind that the CRISPRi mediated gene repression is inducible and fully reversible.
The efficiency of the repression can be altered by incorporating mismatches into the sgRNA where the target sequence will bind. By doing so we also can regulate how much gene repression we want.
Advantages of CRISPRi (interference):
Now one may have a question: if we have RNA interference like advanced techniques, why do we need to develop tedious and costlier CRISPR interference for gene repression?
The main reason is off-targeting.
The RNAi shows more off-targeting effects than the CRISPRi. The CRISPRi mediated transcription repression is far more gene-specific than the RNAi.
Besides, the RNAi is complex, low efficient, unpredictable and mRNA specific. Take a look at the major differences between both techniques.
| RNAi | CRISPRi |
| Repression at mRNA level | Repression at transcription level- initiation and elongation |
| Less effective | More effective |
| Less efficient | More efficient |
| Off-targeting | target-specific |
| Works for selected organisms | Works for prokaryotes and eukaryotes both |
| More complex | Simple and easy to use |
| Less costly | More costly |
The present technique can do partial as well as full repression up to 99.9% with great precision.
The strength of the gene repression can be changed as per the requirement by changing a few or multiple bases from the sgRNA.
It gives more options for optimization and customization as per the requirement of the assay.
The present is independent, straightforward and direct as it depends on Watson and Crick base pairing between the sgRNA and target sequence.
The present technique targets at the DNA level (during transcription) but not on the transcript (mRNA) and hence it doesn’t compete with cells’ natural endogenous mechanism like the RNAi.
One of the incredible advantages of the CRISPRi is that multiple genes can be targeted, depending upon multiple gene targets various unique sRNAs are designed and synthesized.
Limitations of CRISPRi (interference):
One of the biggest setbacks of the present technology is that it relies on the PAM- protospacer adjacent motif sequence in the target gene. Meaning, the CAS every time requires PAM to recognize.
The sgRNA has only 14 nucleotides complementary to the target loci which is too short and increases the chance of non-specific or off-target binding.
Besides these two major limitations, the present technique is costly and tedious to perform including steps of CAS designing, sgRNA designing and synthesis etc.
Applications of CRISPRi (interference):
The sky is the limit for CRISPR and associated technologies as of now. I have listed tons of applications of CRISPR-CAS9 in our previous article, first read it here:
Now coming to the present topic, CRISPRi (interference) based gene repression can be applied in gene knockdown, genomic loci imaging, stem cell research and genetic screening.
Gene knockdown by CRISPRi is used for prokaryotic and eukaryotic gene silencing; off-target analysis, leaky and partial repression studies on E.coli and B. subtilis are now ongoing.
It’s a robust tool for investigating gene functions and other cellular processes.
It has also been used in metabolic engineering.
It has also been applied to attenuated virulence genes in bacteria and other pathogens.
It also provides novel gene targets for antibiotic and vaccine development.
Recent studies also suggest that phenotypes of prokaryote essential genes are studied. In addition to this, phenotypic effects of the genes related to pathogenic growth and cell morphology can also be investigated.
The dCAS mediated loci imagining is also more beneficial than FISH in which fluoro- molecule or protein linked with the dCAS9 has been targeted for genomic loci study and imagining.
This allows scientists to study chromatin remodeling, histone modifications and tracking chromosome loci.
Note that every potential application of the CRISPRi listed here relies on gene knockdown, not on gene knockout.
Wrapping up:
As I say in every CRISPR article, CRISPR is amazing, by far it’s the technique superior to any other available gene-editing tools. The reason is its infinite uses and future applications.
However, it is yet not ready to use mankind’s until its potential risks and setbacks are ruled out. But still, it’s a future, a future of genetics and of the whole world.
Keep linked with our blog to learn more on these topics, more amazing topics will be covered soon, till then please read out the previous article and do Bookmark our website.
Read more: The Curious Case of CRISPR-Babies.