“The sgRNA or gRNA is known as single-stranded guided RNA or guided RNA used in the gene-editing technique which makes a complex with the CAS9 protein to cleave the DNA.”
The Gene editing process is employed to edit, insert or delete genes from a genome aiming to alter someone’s DNA to improve health.
Various gene editing or genome editing techniques are now available in which CRISPR-CAS9 is widely used.
CRISPR-CAS9 is a bacterial defense system used to edit genes at a specific location. Here the CRISPR is nucleic acid sequences and CAS9 is a nuclease protein that uses the CRISPR sequences to cleave DNA.
Using this mechanism Bacteria invades pathogenic attacks. Scientists are utilizing this system for the targeted sequence DNA cleaving. CAS9 is an excellent tool to do so. But without the use of guided RNA or sgRNA, the CAS9 can’t find its target location in a genome.
So how does the guided RNA guide nuclease to work in gene editing?
In the present article, I will try to give you an insight into the whole mechanism mediated by the sgRNA and how we can design it in silico.
Related article: How does the CRISPR CAS9 System Distinguish Self vs Nonself DNA?- 7 Proven mechanisms.
Key Topics:
What is sgRNA?
The first sgRNA was discovered by Bakalara N, Blum B and L Simpson from the mitochondria. The first sgRNA discovered by the trios has the power to delete, insert and cleave DNA within the prokaryotic system.
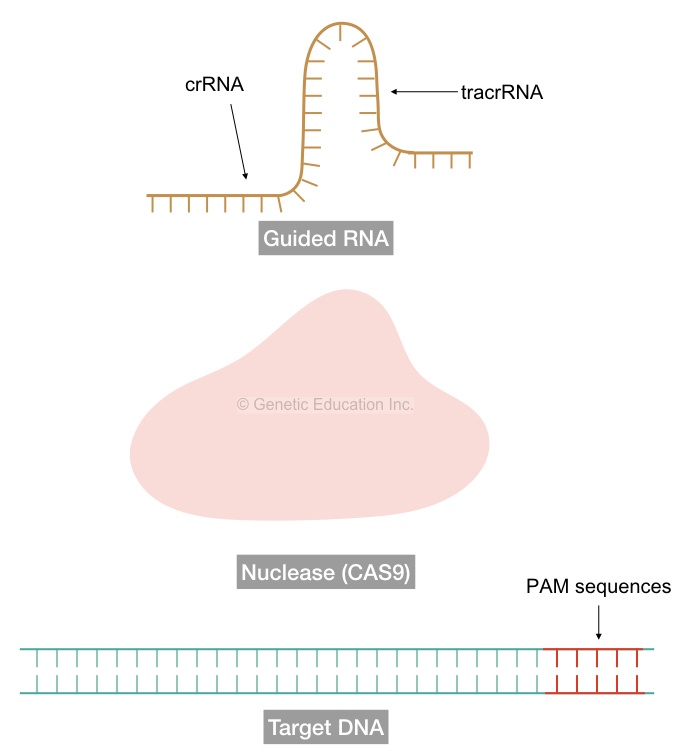
sgRNAs are the sequences of RNA molecules that bind to the target location and thereby facilitate binding of nuclease- the case CA9 to the target location to cleave.
To perform this function efficiently, the single-stranded guided RNA (sgRNA) has two different sequences in it.
The first sequences are specific to the CRISPR or the gene of our interest (for in vivo experiments) known as crRNA of 17-20 nucleotides and the rest of the sequences are of nuclease specific known as tarcrRNAs. The tracrRNA facilitates the binding of nuclease to the sgRNA.
The confusion…
The field of gene editing, especially associated with CRISPR-CAS9 is new and revolutionary obviously, but though the terms gRNA and sgRNA are considered the same, some scientists consider them as different.
Specifically talking, the sgRNA is made up of crRNA and trcrRNA but the guided RNA is only a single unit that guides the nuclease activity.
Anyway, by using artificial crRNAs for a gene of our interest and combining it with the gRNA, we can cleave DNA at a specific location.
Here in the present article, we will consider gRNA and sgRNA as the same to reduce the complexity of the understanding.
In the prokaryotic system:
The first gRNA and CRISPR-CAS9 are discovered from prokaryotes. It is present in mitochondria in which it performs the function of RNA editing to correct the errors.
The gRNAs are smaller, non-coding and single-stranded RNA guide prokaryotic nucleases to repair the errors in 3’ to 5’ direction.
Definition:
“The sgRNA- single-stranded guided RNA or guided RNA is a type of nucleic acid sequence used in gene editing for target-specific cleavage.
Properties of sgRNA:
The single-stranded guided RNA should have several properties that must be present when we design the sgRNA.
First, one of the important characteristics a guided RNA should have is the presence of crRNA and tracrRNA sequences to bind with the target sequence and nuclease domain, respectively. Besides, several other properties are,
- It should be single-stranded.
- It should be 17 to 25 nucleotides long, lower gRNA slips target-specificity.
- To stabilize the DNA-RNA complex, the gRNA should have a high GC content, approximately 40 to 80%.
- The gRNA shouldn’t be too short, otherwise, the binding will not happen.
- The crRNA part should be 20 nucleotides long, approximately.
Designing the sgRNA:
To perform various gene editing, different sgRNAs are designed to target different genes or DNA sequences we wish to edit. Based upon the sequence information the crRNA and tracrRNA are designed.
(We know the function of both!)
In the very first step, identify the PAM sequence in our gene of interest. The PAM- sequence is the proto-spacer adjacent motif on DNA. Above or upstream to the PAM region is our target location.
Design the crRNA 20 nucleotides upstream to the PAM. but remember, it should have several unique features. We have listed it in the above section.
The PAM sequence should not be included in a sgRNA construction. However, to locate and perform cleaving for the nuclease, the PAM sequences must be required.
We can select the target site on either strand.
In the last step, the tracrRNA is designed which should have the sequences that can fold and make a hairpin-like loop and bind to each other.
The tracrRNA loop is first recognized by the CAS9 nuclease, the sequence of the crRNA (the single-stranded one, what we have designed so far!) leads to the target location.
Finally, the nuclease identifies the target location and cleaves.
However, designing the sgRNA manually is a tedious and time-consuming process. Using online tools can help to save time. Several tools are listed here:
Mechanism of action:
The sgRNA or gRNA mediated gene-editing system is mostly used in combination with the CRISPR-CAS9. The Streptococcus pyogenes CRISPR-CAS9 system is widely used in research, gene therapy and gene editing.
Here it required three parts: the CRISPR sequences that is the crRNA, the nuclease binding RNA that is the tracrRNA (combinedly known as the gRNA) and the nuclease CAS9 itself, obviously.
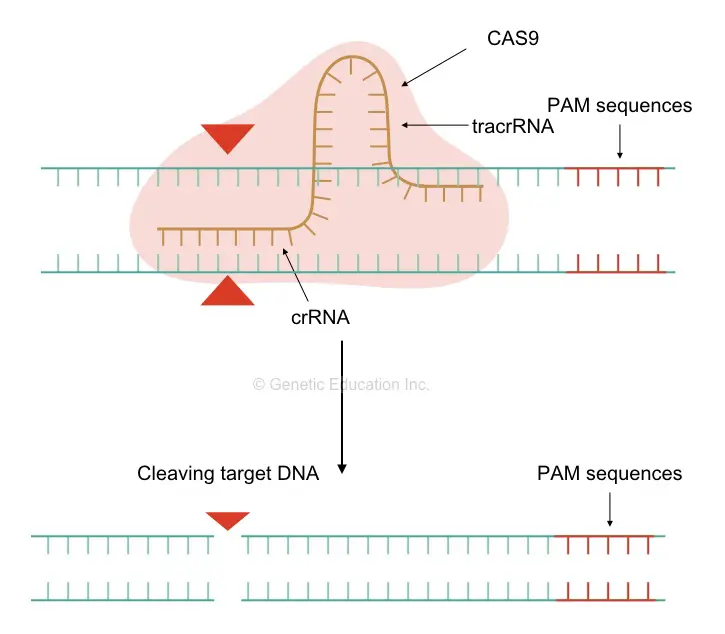
The mechanism of its action is simply clear to us now. A sgRNA’s tracrRNA first recognizes the CAS9 and binds to it. The CAS9 gets sequence information from the crRNA and moves to the target location.
Once it finds it, the crRNA first binds to the target location. Because of the high GC-rich region of the crRNA and the long single-stranded end it stabilizes the DNA-RNA complex.
Once the binding of DNA: RNA completes, the CAS9 starts the cleaving activity. First, it identifies the PAM sequence and starts cleaving upstream to it at where the crRNA is located. Here, it is very essential to understand what a PAM sequence is.
Read more: Cas9 Protein: Structure, Function, Types and Importance.
What is the PAM sequence?
The protospacer adjacent motif is the region in the genome where our nuclease will bind to perform its activity. It is simply, 5’-NGG-3’ region (where N is A, T, G or C). The PAM sequences are actually the DNA region on the target DNA strand, where the nuclease will settle. Different nucleases have different PAM location sequences but the PAM sequence will never take part in the editing, though, very crucial.
Advantages of sgRNA:
It is an efficient, accurate, and cost-effective model for gene editing.
No cloning, sequencing, microarray or in vitro transcription is needed.
In vitro chemical, modifications can be performed.
The editing efficiency of the sgRNA mediated CRISPR-CAS9 system is almost 97%.
Validating the sgRNA:
Once the gene-editing experiment gets over, it is very necessary to validate whether it worked properly or not. There are a few methods we can use to detect, test or validate the sgRNA.
The first method to test sgRNA is the use of an in vitro gene expression system. In which first, the sgRNA is synthesized and transcribed using the transcription kit and purified.
Soon after, perform the editing and run on a gel with negative control and positive control.
The negative control isn’t treated with the nuclease while the positive control is the correct validated results. If we get two bands in our sample, the sgRNA works properly otherwise we have to repeat the experiments.
Another method can detect the sgRNA directly from the cell line. Once the editing is done, PCR is performed in order to validate the results.
Yet another proven method is to use a model organism. The gene-editing mediated by the sgRNA is directly done in a cell line, the embryo is grown and DNA sequencing is performed to know the results.
Related article: What is gene therapy? and How does it works?
Conclusion:
Among all gene-editing systems, the CRISPR-CAS9 proved to be a standout winner with high efficiency and accuracy. Single-stranded guided RNA is a powerful tool that facilitates CAS9 nuclease, a great power to cut DNA at a target location.