“CRISPR works magically in every sense! Every time scientists doubt something and do research, they get more than that, something new and amazing. Yet, it’s not so complex.”
Do you know?
- The total size of phages on earth is 10 billion trillion trillion which makes them the most abundant biological organism on earth.
- The average genome size of a bacteriophage is 35 to 100 Kb while the average size of the bacterial genome is 5bp.
- Phages continuously attack bacteria, interestingly there are 10 phages per bacterium.
Every organism on earth has a well-equipped and powerful self-defense system. Vertebrates have complex and well-studied systems while prokaryotes (bacteria) have a simple but effective immune system.
However, either system has a strong immune response and powerful components to protect its host.
Specifically, talking about bacteria, phages have continuously attacked bacteria since evolution, continue till today and will attack forever because it’s their reason to survive on earth.
Until the discovery of CRISPR in bacteria, the whole mechanism of defense remained unclear. CRISPR-mediated adaptive immunity is a simple mechanism in which a piece of invader DNA is stored and used as a memory unit against future attacks.
Isn’t it fascinating!
In the process, the nuclease activity cleaves off phage DNA and incorporates it into the bacterial genome at a place known as the ‘CRISPR array’ or ‘CRISPR locus’. The same sequence is used as a unit of memory to destroy the same DNA-derived phage in the future.
The whole process has been discussed in our previous article.
The sequence (of the foreign invader) which is now a part of the bacterial genome is known as a ‘spacer’ and is complementary to the region of phage DNA which is known as ‘protospacer’.
Once the Cas nuclease finds the protospacer region using the spacer, it immediately starts destroying it through a catalytic reaction.
But the question is if both protospacer and spacer sequences are ‘similar type’, how does the bacterial immune system (CRISPR) recognize non-self DNA from self?
Meaning why it can’t cut the spacer but only the protospacer DNA?
If you are on this web page, you probably need a specific answer to this question. It shows how far you go into the CRISPR topic and you studied the topic way more than any other research aspirant. My mind raised the same question as yours.
How bacteria distinguish host vs invading DNA to destroy, has been indeed hard to answer question, the reason might be understood to us; it’s a part of an adaptive immune system having a well-developed, diverse and precise non-self recognition system.
In addition, it has evolved, changed and adapted timely to avoid autoimmunity and self-destruction. There are a couple of hypotheses and a few proven theories and pathways that describe the way the bacterial immune system finds non-self DNA.
So let us understand 5 ways using which bacteria distinguish self vs non-self DNA.
You can read many more articles on CRISPR through this category: CRISPR-CAS9.
Key Topics:
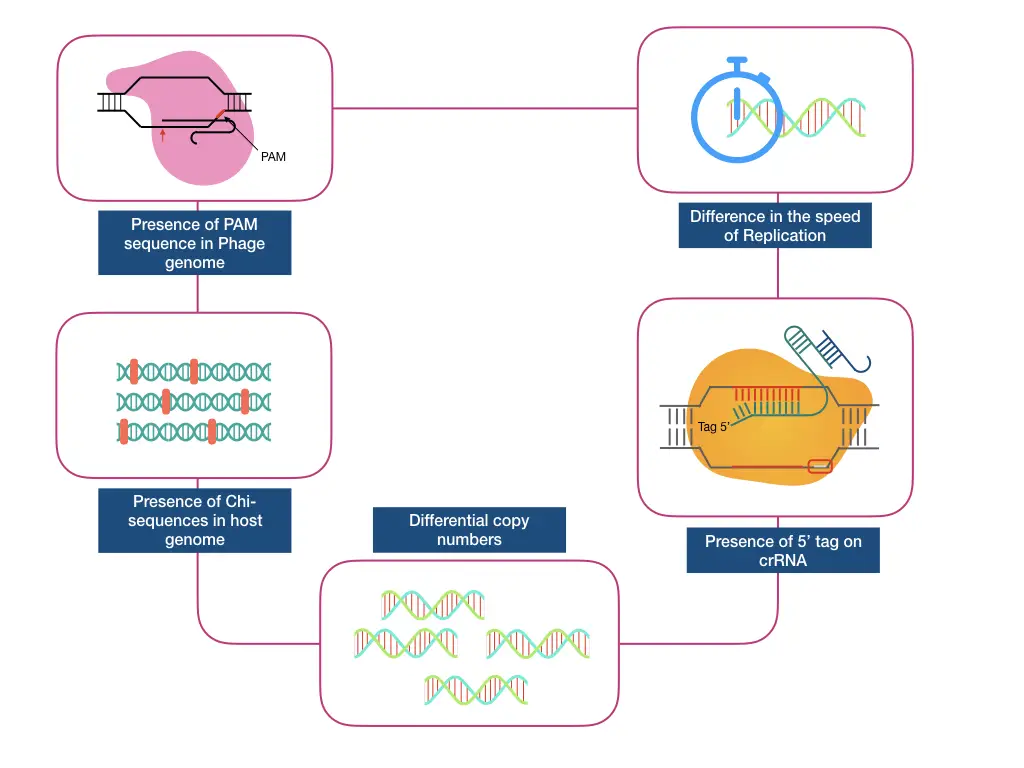
7 ways using which bacteria distinguish self vs non-self DNA:
- The difference in the speed of replication between host and invading DNA.
- Presence of “Chi-sequences” in the host genome
- Presence of unique 5’ Tag in type III system
- Presence of PAM sequence on the invading DNA
- Presence of Restriction modification system along with CRISPR:
- The number of copies of phage or plasmid
- Presence of PFS
Before going further, it’s better to build a strong topic background. Bacteria executes the cause of phage destruction in three distinct steps: spacer acquisition, crRNA maturation and interference.
- Cas2 and Cas1 mediated spacer acquisition cuts the foreign DNA and incorporates it into the host CRISPR locus.
- A cell manufactures crRNA from the spacer by transcription and prepares it for Cas9 to interfere.
- Cas9 mediated interference allows migration of crRNA and Cas9 complex to the target site and cleaves off it.
Spacer acquisition and interference are two steps during which the bacterium mostly distinguishes self vs non-self DNA using several ways.
The difference in the speed of replication between host and invading DNA:
Qimron S et al., performed experiments to validate how the self vs nonself distinguishing mechanism works. Experiments suggest that the Cas1 and Cas2 protein, involved in the spacer acquisition, finds rapidly replicating DNA, located after the replication termination site; mostly. This rapidly replicating DNA is the invading DNA, identified by the Cas1 and Cas2 for further acquisition.
Put simply, their findings conclude that the invading DNA replicates at a faster speed in comparison to the bacterial own DNA. Notwithstanding, they also concluded that speed of replication isn’t the only factor involved in this very precise mechanism.
The presence of Chi-sequences:
Yet another study demonstrated that a specialized region or sequence known as the “chi-sequences” helps distinguish self vs non-self DNA.
The chi sequences work as a marker region, present only on the bacterial own DNA, very frequently; that disallows the CRISPR system to work when present. Meaning, Cas protein only works on “chi-less” sequences, which are the viral/phage DNA sequences.
The presence of Chi-sequences often in bacteria’s genome, though adds an extra layer to their ‘self-security’ but not the only reliable mechanism much like the speed or replication.
Regions on the phage DNA also work as a marker for non-self destruction, greatly decreasing the tendency to autoimmunity, for example, the PAM sequence.
Presence of PAM in invading genome:
Recent advancements in the CRISPR research explain that the presence of PAM- protospacer Adjacent motif present near the RNA-DNA junction, at a target location has a pivotal role here.
PAM sequence has been characterized as 2 to ~6 nucleotides long marker region on the target sequence (or phage DNA) which the crRNA-Cas complex identifies to distinguish between self vs non-self DNA.
PAM is located upstream or downstream of the protospacer, depending upon the type of CRISPR system. The RNA-Cas complex first recognizes it to settle immediately after the protospacer region (Newsom, 2021). Note that only CRISPR type I, II and V follow the PAM-mediated process.
Nonetheless, recent research provides evidence that mutation within the PAM region disturbs the sense of recognition.
The presence of 5’ tag in type III CRISPR system:
Contrary to PAM-mediated non-self DNA recognition, in the CRISPR type III system, a 5’ short tag present on the mature crRNA helps to do the job. The mature crRNA has a specialized 5’ sequence or tag which doesn’t take part in base pairing with the target sequence.
When the nuclease starts recognizing and finds non-base pairing, it immediately identifies it as a foreign nucleic acid and starts degrading it. Note that only the type III system has a 5’ tag mechanism (Charpetiner E, 2016).
The number of copies of phage or plasmid:
The experiment of Wissman J et al., (2020) demonstrated that, as the process of spacer acquisition increases, the number of plasmid or copies of phage DNA increases in the host cell. Meaning, as the number of copies of plasmid increases, the host distinguishes it as non-self DNA and destroys it.
However, the present finding unintentionally postulates that the CRISPR immune system remains ineffective against those who- replicate at a slow rate, having a single or a few replication forks and those who follow a rolling circle mode of replication.
The present study also favors one of the important evolutionary events that why the CRISPR mediated immune system is only present to some prokaryotes and not all.
The reason is clear, the system evolved in such a way that it only supports systems with a slow replication rate, just like some bacteria or sea archaea. In a rapidly dividing organism, it induces strong autoimmune responses, henceforth, aren’t present in eukaryotes.
Other prokaryotes and bacteria also do not have CRISPR like immune systems too.
Presence of Restriction modification system along with CRISPR:
Restriction Modification also supports the CRISPR-mediated adaptive immune system to identify non-self DNA. Restriction digestion or Restriction modification system is yet another known defense mechanism studied in bacteria.
In this, a restriction enzyme binds to its recognition site and cleaves it, meaning it finds the foreign DNA and digests it, which eventually boosts up spacer acquisition.
It actually provides the raw material for the CRISPR system to use as a spacer for regularly updating the memory. Methylation helps to distinguish the foreign DNA from the host.
Studies also demonstrate that the presence of methylation on the host DNA is also a sign of evidence for the CRISPR to use it as a marker and find the invading DNA to clean the system (Dupuis, 2013; Oliveira, 2014).
Presence of PFS in type VI system:
For the RNA target, the presence of the PFS- protospacer flanking sequence operates the interference and signals for phage DNA destruction (Gladitzsch, 2019).
Besides this well-studied mechanism other processes separately and along with one of these are also preventing autoimmune events.
Related articles:
Wrapping up:
The mechanism of how CRISPR distinguishes invading DNA is complex, indeed a combination of many factors or mechanisms. A single mechanism sometimes fails to avail self-defense, for example, a single point mutation in the PAM sequence.
A single mutation in a PAM entirely lacks the sense for Cas to recognize the target, which is perhaps an easy task for the phage.
Though different systems have separate and well-distinguished non-self recognition systems, each mechanism is backed by other ones.
Sources:
- Dupuis, Marie-Ève et al. “CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance.” Nature communications vol. 4 (2013): 2087. doi:10.1038/ncomms3087.
- Oliveira, Pedro H et al. “The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts.” Nucleic acids research vol. 42,16 (2014): 10618-31. doi:10.1093/nar/gku734.
- Newsom, Sydney et al. “The CRISPR-Cas Mechanism for Adaptive Immunity and Alternate Bacterial Functions Fuels Diverse Biotechnologies.” Frontiers in cellular and infection microbiology vol. 10 619763. 28 Jan. 2021, doi:10.3389/fcimb.2020.619763.
- Hille, Frank, and Emmanuelle Charpentier. “CRISPR-Cas: biology, mechanisms and relevance.” Philosophical transactions of the Royal Society of London. Series B, Biological sciences vol. 371,1707 (2016): 20150496. doi:10.1098/rstb.2015.0496.
- Attar, N. A CRISPR sense of self. Nat Rev Microbiol 13, 329 (2015). https://doi.org/10.1038/nrmicro3498.
—————- End of the article —————-
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.