“dCAS9 is a deactivated or dead Cas protein that lacks nucleophilic activity or simply is a deactivated nuclease CAS9.”
Read more: CRISPR-Cpf1(Cas12a) Gene-Editing System- Explained.
You may wonder,
Without nucleophilic catalytic activity, what is the use of CAS9? Because the ultimate function of the CAS protein is to cut DNA in the CRISPR-CAS9 system, and that’s why it’s there!
You may have this question in your mind by reading the hook line. But let me tell you that it has a significant role as a research tool and exactly “how” that we will discuss in this article.
CRISPR-CAS9 is a system of gene editing (precisely, the precise/specific gene-editing tool) having lucrative applications in medical research, diagnostics, animal research, plant and agriculture science.
Related article:
Indeed, it’s a whole technology that broadly relies on three elements; CRISPR, CAS and gRNA, to learn more on how it’s work you can read this article:
Basically, it’s a process that can edit, manipulate, alter or change the target gene sequence, the entire process, stepwise is explained in this article:
The present technology has actually infinite potential applications and scientists are doing it, expanding its use in diverse fields, for example, manipulating or controlling gene expressions.
Yes, it’s possible now!
So many things are going on around CRISPR, it can even store the data of how cells behave during their whole life and so used as a CAMERA to monitor cells activity.
There are different versions of either CAS, CRISPR or gRNA utilized for various purposes. In the present article, I will explain to you, how one of the most potential variations of the CAS- the dCAS9 is used.
This article contains content on the structure of dCAS, how it’s made and its applications. Hope this article will add tremendous value to your knowledge.
Stay tuned,
Key Topics:
What is dCAS9?
The dCAS9, acronym as “dead” CAS or dead CAS9 is a catalytically inactive protein. The native CAS9 protein is a nuclease that has the power to cut either double-strand or single-strand DNA.
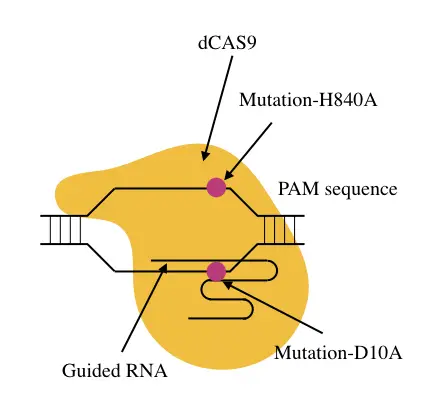
When it combines with CRISPR and gRNA, it specifically cleaves DNA at a programmed location (at target sequence). The catalytic domain of the CAS which has nucleophilic activities makes it possible through rupturing the phosphodiester bonds.
dCAS9 is a specialized, artificially synthesized CAS protein that lacks one of the important properties which is catalytic activity.
The catalytic domain of the enzyme is modified in such a way that it keeps intact its DNA binding abilities but doesn’t execute cleaving. So now after the CAS converting into the dCAS, it has been applied to deliver any functional domain or protein to a specific DNA location.
“deCASing” can be achieved by introducing a mutation into the catalytic domain either D10A or H840A. Either mutation is designed for only preventing enzymatic activity but not DNA binding capability.
That DNA binding capability of the dead CAS, scientists use to perform various functions like altering epigenetic activities, investigating chromatin structure, base editing, epigenetic remodeling and programming chromatin interactions.
Related article:
Concept of dCAS9
Two functional domains of the native CAS endonuclease protein are RuvC and HNH domains that are certainly involved in the DNA cleaving activity; however, domains other than RuvC and HNH have DNA binding activities, perhaps.
(I don’t know exactly what they are!)
When a point mutation either D10A or H849A is inserted into either domain, it becomes catalytically inactive but still, DNA domains do their work finely that are utilized for various purposes.
Functions of dCAS9
dCAS9 performs many separate functions some common functions are;
- Epigenetic remodeling
- Suppressing or activating gene expression
- Base editing
- Chromatin remodeling
- Studying chromatin interaction by programmed dCAS
- Transcriptional regulation
Now let us discuss each function separately.
Epigenetic remodeling:
Epigenetics broadly is a heritable genetic characteristic that is a cell-specific gene expression and regulation achieved by the interaction between DNA and transcriptional factors.
Change in the epigenetic profile of a cell definitely links the condition with some disease or abnormality. Common epigenetic factors are DNA methylations, histone modifications, chromatin remodeling and other post-transcriptional modifications.
Recently technological advancements like the dCAS9 makes it possible to regulate or remodel cells’ epigenetics.
For example,
- Change in DNA methylation or demethylation
- Histone acetylation or deacetylation
- Histone methylation or demethylation
- The dCAS can change any/specific epigenetic factor to achieve gene expression.
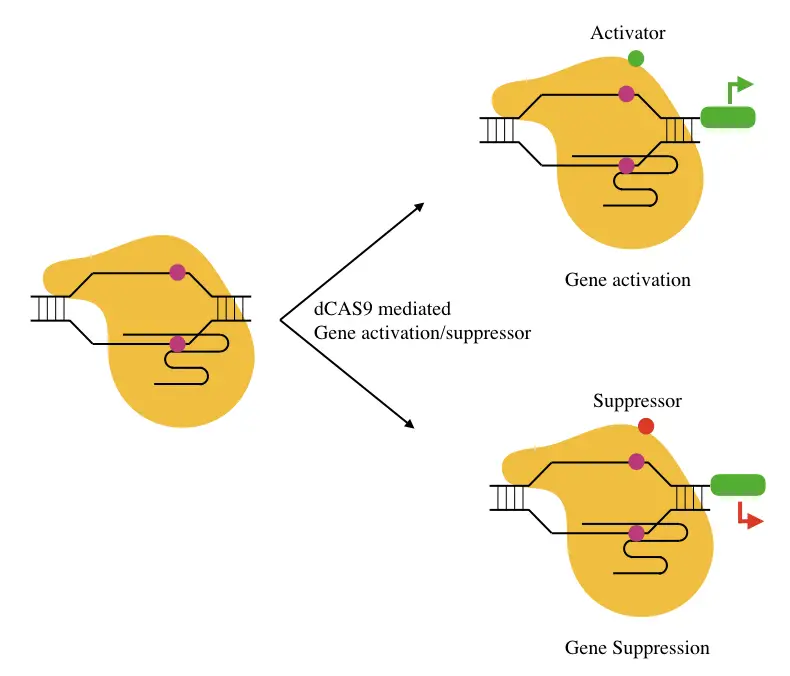
Activating or suppression gene expression:
Transcriptional factors, activators, promoters, enhancers and repressors are several types of proteins that decide the activity of a gene. For example, the presence of the activators on the promoter regions states that a gene is going to be expressed.
Likewise, when repressors present along with the transcriptional factors at promoter regions certainly inactivates gene expression.
A specific mutant dCAS9 when binds to a gene can possibly block the binding of either activator or enhancer, meaning, it prevents a gene to be expressed. So ideally, if a gene is cancerous, its activity can be managed.
The same principle applies to promote gene expression when the dCAS binds to the location for gene repressor.
Scientists also have designed a dCAS variant that provides a binding site for either an activator or an enhancer, and in this way, it also promotes gene expression.
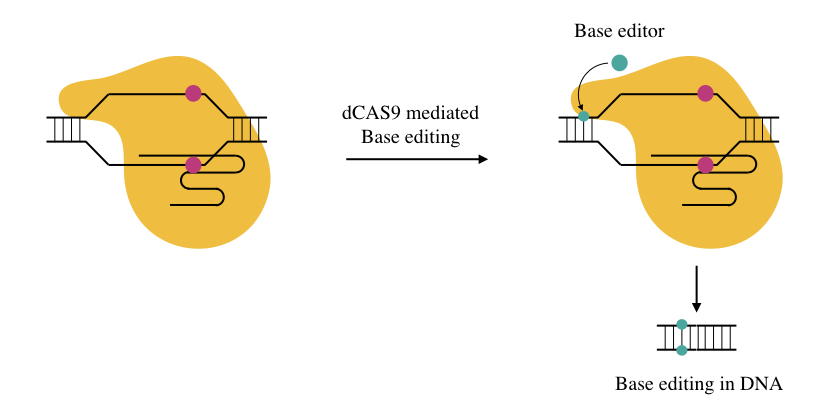
Base editing:
We can modify either DNA or RNA sequences by altering the activity of the base editing enzymes, so we can say it’s a new age genome editing in which instead of doing a single or double-stranded break into the DNA, a point mutation can directly be introduced into the sequence.
Why do scientists do this?
Sometimes dsDNA breaks can’t be repaired efficiently by cells’ repair enzymes, and it ultimately fails the whole experiment. Base editors like cytidine deaminases or adenosine deaminases when linked with dCAS to a specific location effectively changes a single base.
The technique has even more potential applications.
It can even repair mutations! Yes, you hear it right.
A point mutation, if is pathogenic and should be fixed, can be replaced by the correct base by base editing using the dCAS9.
Chromatin remodeling:
The dCAS9-like tools enable scientists to investigate and study chromatin structure. The activity of a gene is determined by the interaction between the DNA, proteins and associated RNAs.
It determines the boundaries of euchromatin and heterochromatin, meaning the transcriptional active and inactive regions. It’s important to note that the ChIP technique can track the remodeling activities but only for predefined and previously studied transcriptional factors.
Note that the mechanism of investigating chromatin remodeling studies is a bit complicated and yet under trial phase but it has the potential to identify previously unknown factors and newer factors involved in the remodeling.
I know some of the tools like the CAPTURE, CasID and CASPEX which are utilized for the study.
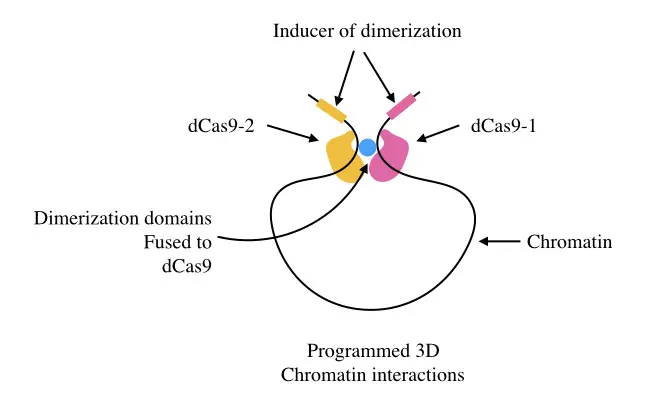
Studying chromatin interaction by programmed dCAS:
The process of investigating the 3D structure of chromatin interaction can be conducted using the dCAS system as well. Here two different CAS9 proteins are used to target genomic regions.
When both interact the structure can be studied in real-time.
Transcriptional regulation:
As we discussed when a programed/synthesized dCAS is linked with either activator or repressor helps to investigate transcriptional activities of a gene.
Applications of dCAS9
As we talked there are numerous uses and applications of dCAS9, here I am enlisting some of the great applications of the present technology.
Altering the deaminase protein domain helps in developing new therapies and manufacturing disease models.
Gehrke J & Cervantes O et al., (2018) and Liang & Ding et al., (2017) introduced a mutation and altered the APOBEC3A and rAPOBEC1 domain to repair the Beta-thalassemia associated mutations.
Similarly, Villiger L and Grisch-Chang H et al., changed the rAPOBEC1 domain to repair the mutation linked with the phenylketonuria.
Zafra M and Schatoff E et al., (2018) altered the same rAPOBEC1 domain for cancer modeling, specifically mutating Ctnnb1 or APC-like genes.
Gaudelli N & Komor A et al., (2017) altered the TadA domain of the deaminase to model the hereditary persistence of fetal hemoglobin syndrome.
Recent studies indicate that several modifications of the dCAS9 have been utilized for treating cancer, genetic disorders, infectious diseases, metabolic and inflammatory disease and stem cell research.
Moses C & Nugent F et al., (2019); Braun C & Bruno P et al., (2016) and Kardooni H & Gonzalez- Gualdna E et al., (2018) utilized dCas9-VP64 and dCas9-VPR for activating some tumor suppressor genes like the PTEN, CHEK2 and DKK3, respectively.
Tumor suppressor genes reduce tumor progression, these tools are under the R & D for treating cancer. Guo J & Ma D et al., (2017) used the same tool dCas9-VPR in stem cell research, especially maintaining the pluripotency by upregulating the NANOG gene.
Variants of dCAS9 tool like the dCas9-VP64, dCas9-VPR, SunTag-VP64 and SunTag-p65-HSF1 are employed in various stem cell research fields (Brezgin S & Kostyusheva A et al., 2019).
Variants of the dCAS tool such as dCas-p300 and Scaffold-MCP-p65-HSF1, scientists are using for treating infectious diseases like HIV and HBV. Zhang Y & Ozono et al., (2019) and Bogerd H & Kornepati A et al., (2015) activated BST2/tetherin, APOBEC3B and APOBEC3G for treating HIV.
Recent studies also show the same tool and set of gene-activation has been applied for HBV therapy as well.
Pinto B & Saxena T et al., (2017) used the dCAS9 tool for blocking the microsatellite repeats in Myotonic dystrophy types 1 and 2.
Kemaladewi D & Bassi P et al., (2019) employed dCas9-VP65 to treat congenital muscular dystrophy type 1A by lifting Lama1 gene expression (upregulation).
Thakore P & Kwon J et al., (2018) reduced serum cholesterol levels by repressing Pcsk9 using the dCAS9-KRAB. The same tool was used by Farhang N & Brunger J et al., (2017); Farhang & Ginley-Hidinger M et al., (2019) and stover J & Farhang N et al., (2019) for repressing TNFR1, IL1R1 and IL6st genes, respectively.
In addition to this, as we said, the present tool is used for transcriptional regulation, epigenetic remodeling, annotating regulatory elements, cell signaling analysis, analyzing gene function and the human genome and identification of antiviral factors.
Tools and variations of dCAS9:
- dCas13b-ADAR2 (RESCUE)
- dCas13b-ADAR2 (REPAIR)
- dCas9-TadA
- dCas9- rAPOBEC1
- dCas9- APOBEC3A
- dCas9-AID
- dCas/MCP-AID
- SunTag-p65-HSF1
- SunTag-VP64
- dCas9-VPR
- dCas9-p300
- dCas9-DNMT3A
- dCas9-KRAB
- dCas9-Ezh2
- dCas9-VP160
- dCas9-KRAB-MeCP2
dCas9-KRAB:
The dCas9-KRAB is one of the most common variations of the dCAS system in which the KRAB is Kruppel- associated box domain. This variant is specifically used for transcriptional repression of genes at endogenous genomic loci.
dCAS9- VPR:
The dCas9-VPR is yet another variant that promotes or upregulates gene regulation. Here the endonuclease that lacks Cas9 is linked with the C-terminal end of transcriptional activators VP64,p65 and Rta.It activates the transcriptional active site of a gene.
dCAS9 TET1:
The dCAS9-TET1 is also used as a gene activator by demethylation. The TET enzyme promotes demethylation, meaning removes the methyl group from the DNA and activates a gene.
dCAS9 mouse:
The dCas9 mouse or knockout mouse is an experimental model organism in which the CRISPR- dCas9 system is used, usually for gene activation/repression, epigenetic modeling and chromatin remodeling studies.
dCAS9 mouse is also used for investigating disease models, cancer study and other disease model studies.
Wrapping up:
CRISPR-CAS9 is an amazing genetic tool, and can change the fate of humanity, perhaps; however, its use should be monitored and regulated. dCAS9 is the variation of the native CAS protein.
The dCAS has so many applications in clinical research as well as in disease studies. I have explained things elaboratively and in layman, so that novice students can also understand them.
I hope you people like this article.
Notedly, please go through the references given below to validate and authenticate the information.
FAQs:
References:
Brezgin S, Kostyusheva A, Kostyushev D, Chulanov V. Dead Cas Systems: Types, Principles, and Applications. Int J Mol Sci. 2019;20(23):6041. Published 2019 Nov 30. doi:10.3390/ijms20236041.
Zafra, M. P., Schatoff, E. M., Katti, A., Foronda, M., Breinig, M., Schweitzer, A. Y., Simon, A., Han, T., Goswami, S., Montgomery, E., Thibado, J., Kastenhuber, E. R., Sánchez-Rivera, F. J., Shi, J., Vakoc, C. R., Lowe, S. W., Tschaharganeh, D. F., & Dow, L. E. (2018). Optimized base editors enable efficient editing in cells, organoids and mice. Nature biotechnology, 36(9), 888–893. https://doi.org/10.1038/nbt.4194.
Liang, P., Ding, C., Sun, H., Xie, X., Xu, Y., Zhang, X., Sun, Y., Xiong, Y., Ma, W., Liu, Y., Wang, Y., Fang, J., Liu, D., Songyang, Z., Zhou, C., & Huang, J. (2017). Correction of β-thalassemia mutant by base editor in human embryos. Protein & cell, 8(11), 811–822. https://doi.org/10.1007/s13238-017-0475-6.
Gehrke, J. M., Cervantes, O., Clement, M. K., Wu, Y., Zeng, J., Bauer, D. E., Pinello, L., & Joung, J. K. (2018). An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nature biotechnology, 36(10), 977–982. https://doi.org/10.1038/nbt.4199.
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., & Liu, D. R. (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551(7681), 464–471. https://doi.org/10.1038/nature24644.
Moses, C., Nugent, F., Waryah, C. B., Garcia-Bloj, B., Harvey, A. R., & Blancafort, P. (2019). Activating PTEN Tumor Suppressor Expression with the CRISPR/dCas9 System. Molecular therapy. Nucleic acids, 14, 287–300. https://doi.org/10.1016/j.omtn.2018.12.003.
Braun, C. J., Bruno, P. M., Horlbeck, M. A., Gilbert, L. A., Weissman, J. S., & Hemann, M. T. (2016). Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proceedings of the National Academy of Sciences of the United States of America, 113(27), E3892–E3900. https://doi.org/10.1073/pnas.1600582113.
Kardooni, H., Gonzalez-Gualda, E., Stylianakis, E., Saffaran, S., Waxman, J., & Kypta, R. M. (2018). CRISPR-Mediated Reactivation of DKK3 Expression Attenuates TGF-β Signaling in Prostate Cancer. Cancers, 10(6), 165. https://doi.org/10.3390/cancers10060165.
Zhang, Y., Ozono, S., Yao, W., Tobiume, M., Yamaoka, S., Kishigami, S., Fujita, H., & Tokunaga, K. (2019). CRISPR-mediated activation of endogenous BST-2/tetherin expression inhibits wild-type HIV-1 production. Scientific reports, 9(1), 3134. https://doi.org/10.1038/s41598-019-40003-z.
Bogerd, H. P., Kornepati, A. V., Marshall, J. B., Kennedy, E. M., & Cullen, B. R. (2015). Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proceedings of the National Academy of Sciences of the United States of America, 112(52), E7249–E7256. https://doi.org/10.1073/pnas.1516305112.
Bogerd, H. P., Kornepati, A. V., Marshall, J. B., Kennedy, E. M., & Cullen, B. R. (2015). Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proceedings of the National Academy of Sciences of the United States of America, 112(52), E7249–E7256. https://doi.org/10.1073/pnas.1516305112.
Pinto, B. S., Saxena, T., Oliveira, R., Méndez-Gómez, H. R., Cleary, J. D., Denes, L. T., McConnell, O., Arboleda, J., Xia, G., Swanson, M. S., & Wang, E. T. (2017). Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9. Molecular cell, 68(3), 479–490.e5. https://doi.org/10.1016/j.molcel.2017.09.033.
Kemaladewi, D. U., Bassi, P. S., Erwood, S., Al-Basha, D., Gawlik, K. I., Lindsay, K., Hyatt, E., Kember, R., Place, K. M., Marks, R. M., Durbeej, M., Prescott, S. A., Ivakine, E. A., & Cohn, R. D. (2019). A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature, 572(7767), 125–130. https://doi.org/10.1038/s41586-019-1430-x.
Thakore, P. I., Kwon, J. B., Nelson, C. E., Rouse, D. C., Gemberling, M. P., Oliver, M. L., & Gersbach, C. A. (2018). RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nature communications, 9(1), 1674. https://doi.org/10.1038/s41467-018-04048-4.
Farhang, N., Brunger, J. M., Stover, J. D., Thakore, P. I., Lawrence, B., Guilak, F., Gersbach, C. A., Setton, L. A., & Bowles, R. D. (2017). * CRISPR-Based Epigenome Editing of Cytokine Receptors for the Promotion of Cell Survival and Tissue Deposition in Inflammatory Environments. Tissue engineering. Part A, 23(15-16), 738–749. https://doi.org/10.1089/ten.TEA.2016.0441.
Farhang, N., Ginley-Hidinger, M., Berrett, K. C., Gertz, J., Lawrence, B., & Bowles, R. D. (2019). Lentiviral CRISPR Epigenome Editing of Inflammatory Receptors as a Gene Therapy Strategy for Disc Degeneration. Human gene therapy, 30(9), 1161–1175. https://doi.org/10.1089/hum.2019.005.
Stover, J. D., Farhang, N., Lawrence, B., & Bowles, R. D. (2019). Multiplex Epigenome Editing of Dorsal Root Ganglion Neuron Receptors Abolishes Redundant Interleukin 6, Tumor Necrosis Factor Alpha, and Interleukin 1β Signaling by the Degenerative Intervertebral Disc. Human gene therapy, 30(9), 1147–1160. https://doi.org/10.1089/hum.2019.032.
Guo, J., Ma, D., Huang, R., Ming, J., Ye, M., Kee, K., Xie, Z., & Na, J. (2017). An inducible CRISPR-ON system for controllable gene activation in human pluripotent stem cells. Protein & cell, 8(5), 379–393. https://doi.org/10.1007/s13238-016-0360-8.