“CRISPR-CAS9 is a revolutionary system of gene editing or genome editing that uses the guided RNA to cleave the DNA at a specific location in a genome.”
Related article:
Editing a gene or DNA for producing the desired phenotype was like a fairytale in the 80s, however, now we can do it. Tones of gene editing tools are now available, some are complicated & random whilst others are simple & precise.
Why are we doing gene editing? Might be the first question that will strike in your mind when you are thinking about gene editing. It allows us to create various genetically modified organisms, helps to treat genetic disorders and creates economically important plant species.
Several ways to do this are inserting, removing/ deleting sequences from the genome of an organism. CRISPR-CAS9 is one such tool, easy to use, highly accurate and precise.
It allows researchers to manipulate the genome of any organisms like animals, plants or microbes. It’s a tool for genetic engineering we can say, but more powerful than any other tools and techniques available.
Put simply, CRISPR is a nucleic acid sequence and CAS9 is an enzyme that works together. It needs a sgRNA or guided RNA to facilitate target-specific manipulation. The first explanation of the CRISPR-CAS9 system was demonstrated by Yoshizumi Ishino and co-workers in 1987.
Related article: What is gene editing and CRISPR-CAS9?
In the present article, we will discuss the entire process of the CRISPR-CAS9 system and how you can perform it in your lab, virtually. However, what we will discuss is totally different from the actual lab work.
Furthermore, we are not discussing the mechanism, instead, I will explain the process stepwise, so that a newbie student can understand it thoroughly. I hope this guide will add value to your knowledge of genetics,
Stay tuned,
Key Topics:
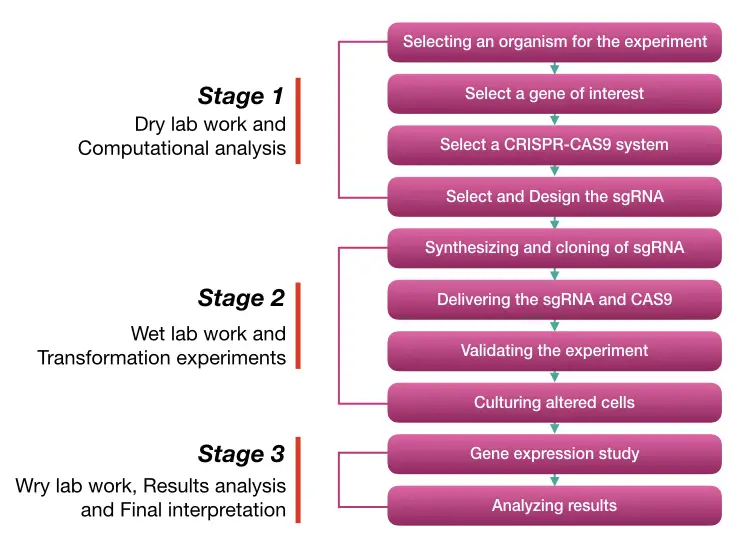
Steps in CRISPR-CAS9:
Several steps to use the CRISPR-CAS9 system for gene editing and genetic engineering are:
- Select an organism for the experiment
- Select a gene of the target location
- Select a CRISPR-CAS9 system
- Select and Design the sgRNA
- Synthesizing and cloning of sgRNA
- Delivering the sgRNA and CAS9
- Validating the experiment
- Culturing the altred cells
- Gene expression study
- Analyzing results
The CRISPR system was first used to study gene knockout in which the gene or DNA sequence is removed from the model organism. Although, in recent trends, it has various applications in various fields.
We will try to explain each step in layman language so that you can understand it well. Instead of going more into scientific writing, I will try to write in a simple language.
Steps and Procedure of CRISPR-CAS9:
Selecting an organism:
The very first step is to select an organism to edit. We can’t perform gene editing for all organisms. Note that no CRISPR-gene therapy is yet approved ‘fully’ to manipulate the human genome.
It can be efficiently used for plants but to use it in treating genetic disorders, first, we have to experiment on model organisms. If we are performing the CRISPR-CAS9 to treat genetic disorders, select the model organism whose genome is closely related to the human.
For example the mice.
Selecting a gene or target location:
Now after selecting the model organism, the gene or the DNA sequence we wish to study or alter or knockout is selected. Suppose we wish to study the IGF gene.
Select that gene, analyze it, research data, observe sequence computationally do other bioinformatics analysis on this gene. Data analysis will allow us to understand whether this sequence can be manipulated or not.
Gather data of the length of the sequence, GC content, phenotype and phenotypic variations, SNPs, other mutations and gene expression. This data will help us to design the guided RNA and locate the PAM sequence to edit a gene.
Now select a region to silence or remove.
Select a CRISPR-CAS9 system:
Not all the CRISPR-CAS9 systems can work for all the manipulations. For example, to perform a knockout experiment we have to select a nickase CAS9. Some of the CAS9 systems are enlisted below
- CAS9 nickase: Gene knockout, change in a sequence
- dCAS9: Activation and repression.
Some other CAS proteins are Cas13, Cas12, Csm, Cmr and RNase III. Different companies have different nomenclature for their own CAS protein.
Conclusively we can say we have to select the CAS9 and CRISPR sequences based on our experimental requirements.
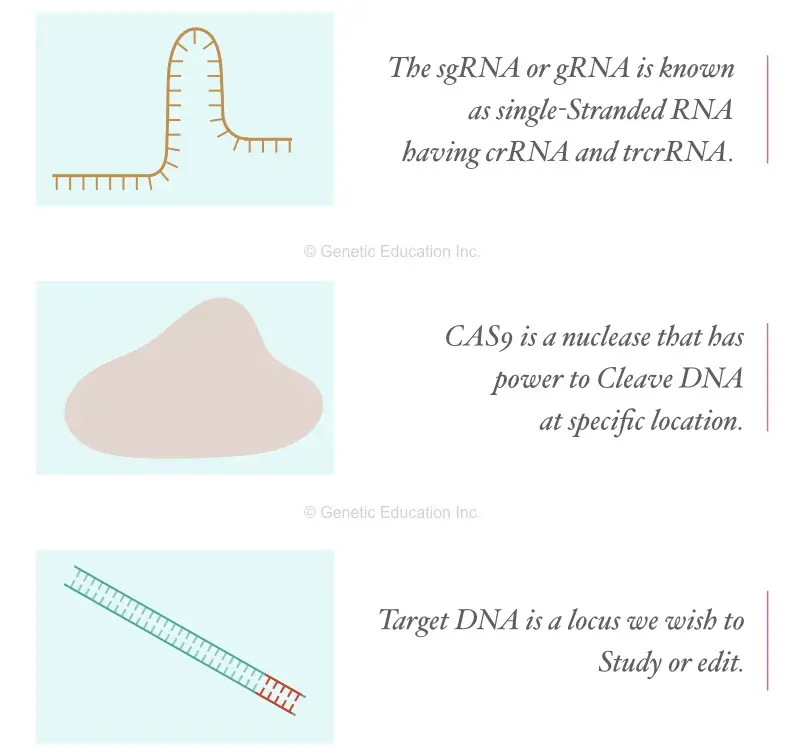
The CAS is a class of protein known as the nuclease that can cleave single-stranded as well as double-stranded DNA.
Selecting and Designing the sgRNA:
sgRNA or gRNA is the short RNA sequence that allows gene editing by targeting a specific location. In the next step, we have to design the synthetic sgRNA, based on the sequence information available.
Shortly I am explaining what sgRNA is!
The sgRNA is a single-stranded RNA known as a guided RNA having a complementary sequence to our target location. It is made up of two parts; the crRNA that has the complementary 20 nucleotides and the tracrRNA having the loop that recognizes the CAS9.
Once the tracrRNA part identifies the CAS, it guides the nucleus to the site of cleaving.
We need to design the sgRNA computationally. First, based on the location of the PAM sequence, the sgRNA binding site is decided, usually, upstream to the PAM.
We have covered an amazing article covering the whole process and tool to design the sgRNA. Read it here: sgRNA- definition, mechanism and designing.
Once the gRNA is designed, we need to synthesize it and clone it in a plasmid.
Synthesizing and cloning of sgRNA:
We now have state-of-the-art facilities to synthesize the oligos of the gRNA or sgRNA in vitro. But we have to prepare a stock or clone of it. To do so, select the specific plasmid, insert the gRNA gene in it and develop several clones of it.
After that, isolate the gRNA expressed from the plasmid. Moreover, In vitro transcription also helps to synthesize the sgRNA. Once our gRNA is synthesized, CAS9 is ready to manipulate the gene of interest.

Delivering the sgRNA and CAS9:
The electroporation method is widely used to insert the CAS9 and sgRNA into the target cell. Here, under the influence of the current, pores are created in a cell that allows the nuclease and sgRNA intake.
Also, instead of CAS sometimes, mRNA specific to it or a gene is inserted to form a CAS9 in a host cell. Because it is very hard to insert a CAS like larger molecules.
Also, viral vectors like adenovirus, Adeno-associated virus, lentivirus and retrovirus are used to perform the same function.
However to perform the transfection-mediated by the viral vector use mRNA specific to the CAS. The virus itself forms the nuclease protein.
Other CRISPR-CAS9 delivery systems are microinjection, gene gun, sonication and chemical modifications.
Now our CAS and sgRNA are in the target cell.
Validating the experiment:
Now it’s time to know whether the knockout occurs accurately or not, we should have to validate our experiment. Three common techniques are polymerase chain reaction, in vitro transcription or DNA sequencing.
Perform DNA sequencing prior to the experiment and after the completion of the experiment, and compare it. It lets us understand whether a gene is successfully removed or not.
PCR validates the experiment by amplifying the target from the modified cells. We have explained the validation method in this section: sgRNA validation methods.
In addition to this, to validate the results we can also perform the restriction digestion experiment by inserting the restriction site at the target location.
Culture the altered cells:
Now we have a cell line of the altered cell which is genetically modified. Culture the cell line in aseptic conditions and using appropriate culture media. Once sufficient amounts of cell lines are obtained, insert them into the target organism or organism we had selected for the experiments.
Note: don’t judge scientifically, it is just a simple explanation of the entire CRISPR-CAS9 mechanism.
How does the gap fill?
We have broadly discussed the process of knockout experiments in which we have removed several sequences. But a gap generated by the CAS9 nuclease activity can’t remain unfilled.
A cell’s DNA repair mechanism repairs the DNA or fills the gap by either Non-homologous end-joining or by direct DNA repair. Here the DNA polymerase fills the gap with nucleotides while the ligase forms the phosphodiester bond to fill the gap.
But, wait for a minute!
The story does not yet end here. We should have to perform several experiments to check the status of our altered cells. Gene expression studies are one among those many options.
Gene expression studies:
We have to check the expression of an altered gene in all cell lines. Suppose we have inserted some DNA sequence, It will give us an idea about whether it is expressed in all cells or not!
RT-PCR or quantitative PCR is performed to check gene expression. Here the mRNA is isolated from the cell line, reverse-transcribed into cDNA, and quantified in a PCR.
If you want to learn more on gene-editing and CRISPR-CAS9, you can read our previous article on this topic:
What is gene editing and CRISPR-CAS9?
Analyzing results:
Results can be analyzed computationally as well as by physical examination. Computational tools help to analyze if everything is completed correctly or not, a gene sequence, gene expression profile etc.
- While the physical examination helps to understand,
- If a new phenotype is induced or not
- If the original phenotype still exists or not,
- If the phenotypic results are wrong or not.
Conclusion:
Briefly, this is the whole process of how a standard CRISPR-CAS9 experiment is conducted in a genetic lab. However, the specificity of the results depends on which system we have selected.
If we select the wrong CAS9, we can’t get the desired results.
SgRNA plays a crucial role in editing as well. If the sgRNA is not designed carefully, It will cleave untargeted regions in a genome. Therefore, which system we are selecting decides what we get!
If you are interested to learn more about the CRISPR-CAS9 system. You can read this article of Addgene: CRISPR guide.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.