“Learn about gene knockout– process, steps and methods, techniques to construct knockout and applications in genetics and clinical research, in this article.”
Did you know that modification in a single gene can have a significant effect on governing traits or phenotypes? Gene knockout is a technique that selectively turns off a specific gene.
Genes are important for us as they encode proteins. These proteins, later on, participate in different cellular activities and metabolic pathways. Studying a gene’s function allows us to understand its role in cellular pathways and how it causes abnormalities.
One incredible technique scientists use is called ‘gene knockout–’ a process to selectively inactivate a gene. Gene knockout is a molecular genetic research technique scientists use to study the function of a gene.
Knocking out allows scientists to understand the importance of a gene by functional analysis. Resultantly, they can understand cellular activities, pathways, metabolic activities and causes of diseases.
In this article, I will take you on an amazing journey in which you can learn about gene knockout– what it is and how it’s done, techniques for knockout and applications in research.
Genetic Education Inc. focuses primarily on conceptual learning. Our team of experienced scientists is dedicated to re-discovering knowledge and presenting it understandably. Let’s learn the key concepts of gene knockout.
Stay tuned.
Related article: How To Construct Knockout Mice? A Stepwise Guide.
Key Topics:
What is gene knockout?
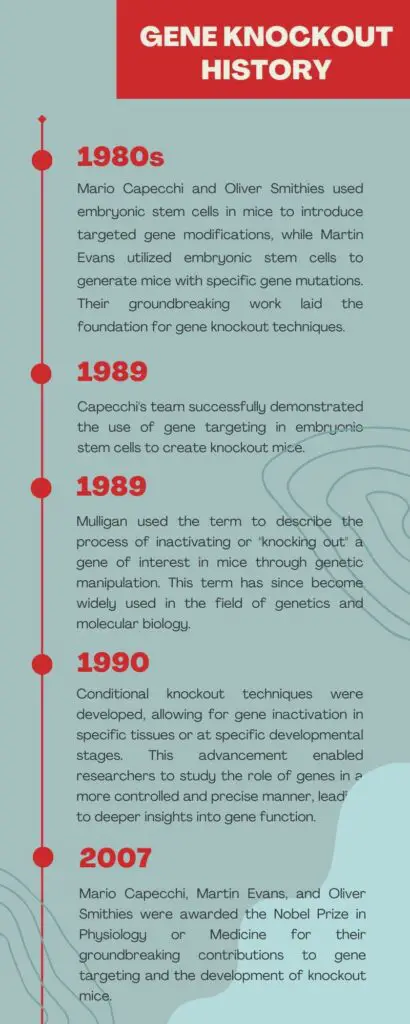
Gene knockout, short for KO, is a molecular genetic technique applied to study a gene’s function. The first successful gene knockout experiment was conducted by Capecchi et al. in 1980.
They disabled a gene from mice embryonic stem cells by homologous recombination technique and the first knockout mouse was born. Capecchi et al. were awarded the Nobel Prize for Physiology and Medicine for their findings.
Some important historical events in gene knockout are listed here in the graphics.
Let’s take an example to understand it better.
MCH gene encodes mouse coat hair (it’s just a hypothetical example). Mice with the MCH gene have shiny, long-coat hair. Suppose we wish to understand the importance of the MCH gene in skin cancer.
We can use the gene knockout technique to inactive the MCH gene and understand its consequences in the progenies. We can introduce a mutation in the coding region, remove one exon, or inactive the promoter region.
By any means, we have to inactive the MCH gene and disallow it to encode the coat hair protein. Using a vector, the altered gene can be introduced in the embryonic cell line. The newly born mice lacking the MCH gene lacks hair on their coat.
However, technically it is known as “naked mice” but we have explained it to understand the concept of KO. That’s how knockout is performed. Remember, the goal here is to suppress or inactivate a gene’s activity selectively.
The mice lacking the MCH gene can be categorized into ‘genetically altered or genetically modified organisms’ and known as ‘knockout mice.’ Now scientists can observe any phenotypic change that occurs by inactivation.
Similarly, the inactivation of two genes is known as a double knockout. Validation assays are performed to prove gene inactivation. This technique is popularly used in microorganism and plant genetic research to conduct functional analysis for economically important traits.
Definition:
“Gene knockout refers to the selective inactivation of a target gene within an organism’s genome using artificial means and to study a gene’s function.”
Or
“To study a gene’s function, a technique that inactivates a gene by artificial mutagenesis or gene manipulation is known as gene knockout.”
By the way, a short form of gene knockout is KO. Now let’s deep dive into the step-by-step process of how this is done.
Process of gene knockout:
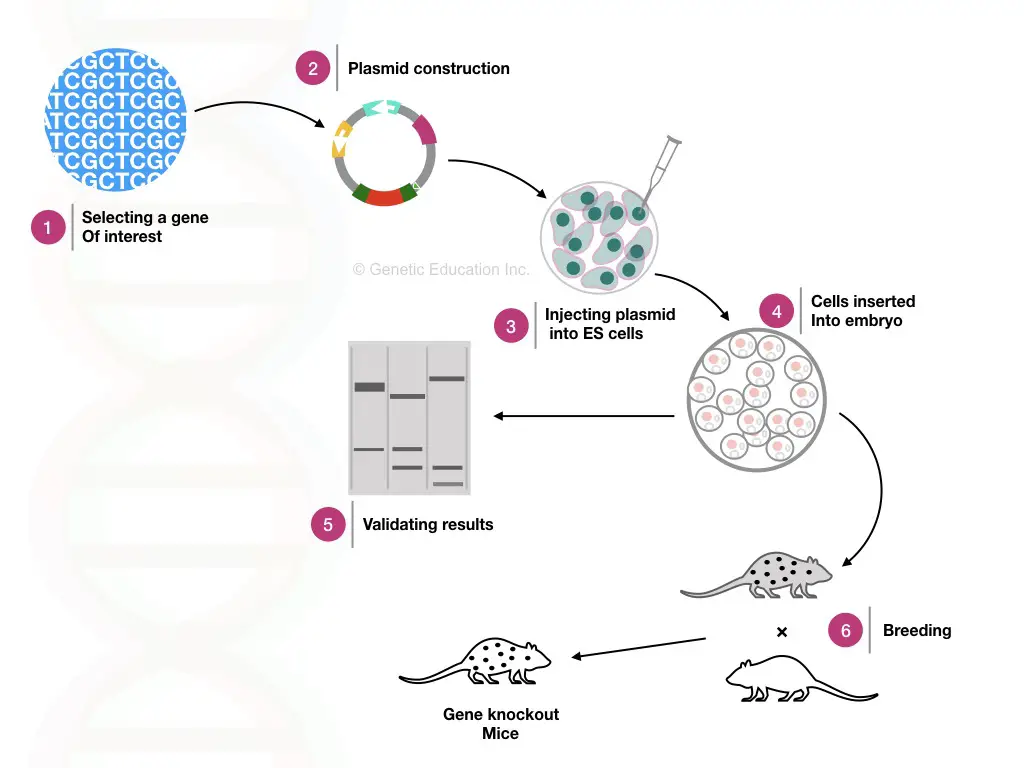
A typical process of KO involves steps like– gene selection, Vector design and construction, transformation, Homologous recombination, selection, identification and validation, insert validation, animal model generation and phenotypic assays.
We can consider gene KO as one kind of genetic engineering experiment.
Gene selection:
The very first step in KO is selecting a gene we want to knock out. A gene is selected only if its function is known and should have been involved in any biological activity or biological process we wish to study.
Some dry lab work has been carried out to understand the structure of a gene. It is mapped on a chromosome and other parameters such as gene length, No of exons and introns, promoter length and sequence, etc are closely examined. Dry lab work helps construct an effective vector for the experiment.
Vector design and construction:
A vector is a vehicle that carries our genetic material and transfers it to another location. Viral vectors, plasmids, cosmids and bacterial artificial chromosomes are a few common types of vectors used in gene therapy experiments.
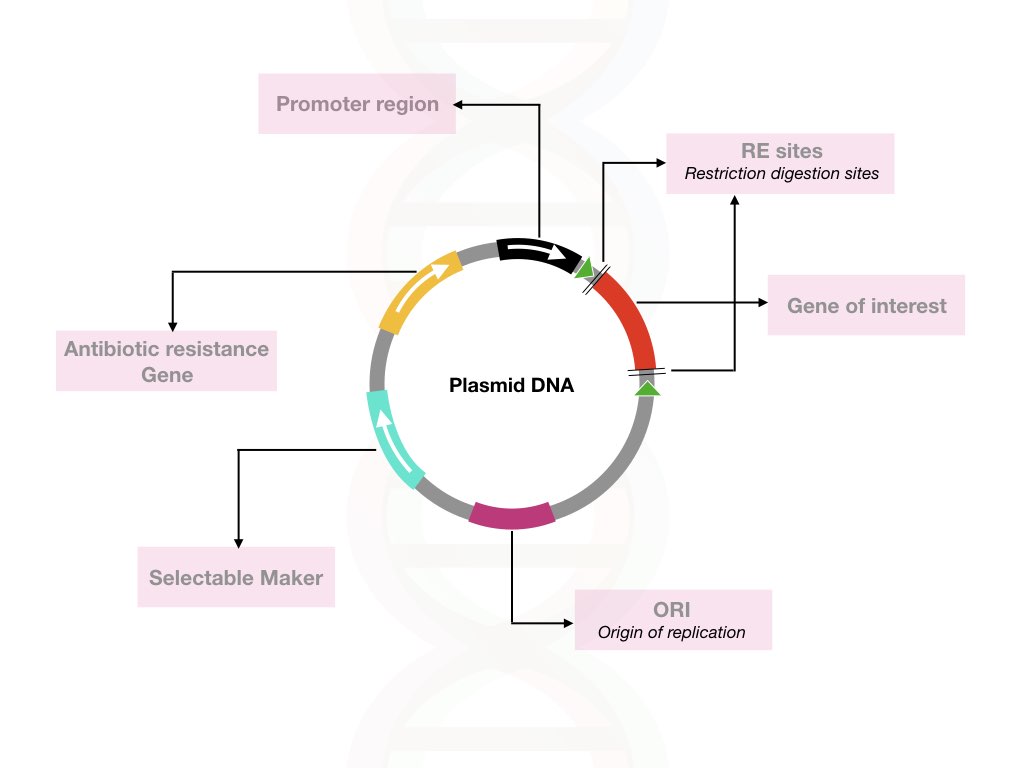
Among all these, a plasmid is the most popular and common vector used in KO. The plasmid is bacterial extrachromosomal and circular DNA that can carry the target mutation or sequence for KO.
Read more on Plasmid DNA: Plasmid DNA: Structure, Function, Isolation and Applications.
A vector consists of the selected marker, a gene-specific homologous region, and a disrupting element- a mutation or sequence. After vector construction and validation, our plasmid vector is ready for transformation.
Note:
It is ideally inserted between the left and right arms or between the target sequence which when recombined with the target genomic region, inserts the marker at its location. Such cells can grow in the presence of Neomycin-containing media.
Transformation:
Now, our plasmid is ready to insert into a target cell. Scientists usually select Embryonic Stem Cells (ESCs) for transformation. ESCs can divide at a faster rate into different cell types and form various tissues.
The vector once finds the gene-specific target location, it recombines and inserts our target ‘change’ with the marker gene in the target cells. Transformed cells are then cultured using an appropriate media.
Electroporation, Sonication and micro-injection are some common techniques for ESCs transformation. Note that cells will only grow if transformation occurs.
Homologous recombination:
After successful transformation, and after reaching the complementary region, homologous recombination will occur which results in the disruption of a gene’s function later on. HR introduces the ‘change’ that eventually knocks out the target gene.
Selection and validation:
Now, it’s important to validate whether the transformation fully occurred or not. Meaning, the experiment is successfully placed or not. Techniques like polymerase chain reaction and DNA sequencing can validate KO.
A few transformed cells are taken for DNA isolation. DNA sequencing can validate KO at the sequence level. PCR and qPCR can also be used for validation using sequence-specific primers.
qPCR can determine the concentration of the target gene from the transformed cells and determines if knockout occurred or not. Besides, restriction digestion can also be employed for KO verification but is less recommended.
After the completion of validation, the successfully transformed cells are selected for the generation of genetically modified model organisms.
Introduction into the animal model:
Now the transformed cells are employed to generate a KO animal using breeding and other techniques.
Phenotypic investigation:
Phenotypic analysis includes physical, biochemical and functional investigations. In vitro, experiments are carried out to study the effect of gene loss. Various chemical assays are also available for protein studies as well.
The entire process of gene knockout is represented in the figure below,
Gene knockout Methods:
So far we have discussed the KO experiment in context with the homologous recombination, however, there are other methods as well that scientists can use. Gene silencing, conditional knockout, gene editing and knockout by mutation are a few common methods for KO. We will discuss each technique one after another in this section. But remember, in any method, the purpose is to inactivate or make silence the target gene viz knockout.
Gene silencing:
Gene silencing aims to deactivate a gene by disrupting its mRNA. A cell’s natural gene regulation mechanism uses RNA interference. Here small complementary microRNAs bind with the mRNA and deactivate them for translation.
During in vitro gene silencing, scientists use artificially synthesized small interfering RNA complementary to the mRNA of the target gene. Upon reaching the target location, it binds with the mRNA, processes it in the RISC complex much like the cell’s natural mechanism and disallows translation.
This process blocks protein translation from the mRNA by mRNA disruption and effectively silences the gene (knockout it). Other techniques like antisense RNAs are also used for gene knockout.
Related articles on RNA interference:
Conditional knockout:
Conditional KO is ‘specific.’ It allows the inactivation of a target gene in a specific tissue or specific developmental stage. Or gene inactivation has been carried out to study a specific function in a specific cell population, tissue or developmental stage.
Conditional KO is a great tool to study tissue-specific cancer. The present technique uses homologous recombination and tissue-specific promoter for conditional KO where knockout only occurs in selected tissues or cells. CKO is the best way to study a gene’s function at a tissue or cellular level.
Interestingly, it is not only performed in the embryonic stage but also on adult animals as well. Cre-LoxP is one such technique in which the “Cre” recombinase works on LoxP tissue-specific inactivation. Recent studies showed that CKO is widely used in breast cancer and ovarian cancer research.
Conditional knockout is very popular and by far the most successful technique for gene inactivation. We will cover the whole topic in some other article.
Gene knockout by mutation:
KO by mutation is yet another important technique. In this technique, a mutation is incorporated in a target gene or sequence in a way that deactivates or alters the target gene’s function.
Such a mutation is known as loss of function mutation which can be either insertion, deletion, duplication or translocation. Artificial mutagenesis techniques like chemical alteration, radiation-induced mutagenesis or physical mutagenesis have been used to incorporate a mutation.
Notedly, artificial mutagenesis induces mutation in a gene’s coding, promoter or any regulatory region to knock it out.
Related article: Site-Directed Mutagenesis- Methods and Applications.
Gene knockout by gene therapy:
Knockout by gene therapy is a novel approach, which involves the delivery of a therapeutic agent, engineered vector or system to suppress a gene’s function.
CRISPR-CAS9 is a popular and modern gene therapy technique that can introduce a mutation or remove a sequence from the target gene. Any target element, for example, the sgRNA in the case of the CRISPR technique, is engineered in a way that introduces a desired change in the DNA sequence.
Such techniques are very accurate and precise for gene knockout. Gene therapy approaches hold promise for potential applications in treating genetic diseases. If you would like to learn more, you can read these articles.
Read more:
- What is Gene Therapy and How Does It Work?
- Gene Therapy: Types, Vector, Process, Applications and Limitations.
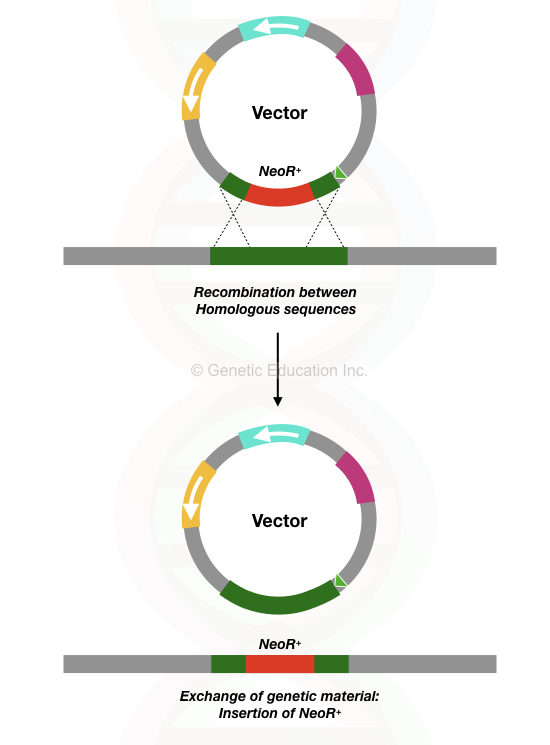
Homologous Recombination:
Homologous recombination involves the exchange of genetic material between two homologous sequences or genes. Homologous recombination has a higher success rate. Thus, Scientists use this phenomenon for KO.
Here, scientists prepare homologous sequences complementary to the flanking region of the target gene and introduce them in the vector. The vector transfers it to the target location, where the recombinase enzyme performs recombination and exchanges the native sequence with our target sequence.
Interestingly, this process either disrupts a gene’s function, eliminates it or suppresses it completely. Notedy, as aforesaid, a marker gene is also incorporated for validation.
Applications of Gene Knockout:
The key function of KO is to eliminate or inactivate a gene. Thus, making it functionally inactive. The present revolutionary approach has opened new doors for researchers to study diseases, genes and traits. Here are some of the amazing and important applications of gene knockout.
Understanding a gene’s function:
Discussion so far demonstrates that the pivotal function of KO is to understand a gene’s function. Scientists can study alterations in phenotypes or traits by selective knockout or target gene inactivation.
KO experiments on model organisms help study genes’ role in development, behavior, biological activities, physiology and disease progression. Eventually, such studies contribute to develop new strategies and therapies for diseases.
Genotype and phenotype relation:
Gene knockout is a great tool to establish the relationship between a gene and a phenotype. Through selective gene inactivation, its effect on a phenotype or group of phenotypes can be studied. This way, scientists understand the relationship between a gene and a phenotype.
Functional genomics:
Functional genomics is largely defined as ‘to understand the overall function of the genome.’ KO is a crucial tool in functional genomic studies. Through selective gene targeting, scientists can systematically investigate the impact of individual genes on related functions, pathways, cellular activities, signaling pathways, etc.
Scientists can understand the function as well as the evolutionary importance of homologous genes and gene families too.
Evolutionary studies:
Gene knockout is also an amazing tool for studying Evolution and the role of a gene in evolutionary events. By selectively inactivating a gene across different organisms, scientists can understand a gene’s effect and its evolutionary importance. This helps to understand the underlying mechanism of evolution and biodiversity.
Disease studies:
KO also helps understand the molecular mechanism behind diseases through gene-level analysis. Scientists prepare animal models to perform KO for a particular disease-governing gene and understand its effect.
These studies help us understand how genetic diseases happen and how certain genes or groups of genes play a role in causing disease. This understanding helps in creating new treatments for the disease.
Drug studies and development:
Assessing a disease at the molecular level and using gene knockout techniques also aids in understanding drug responses. This means that scientists can evaluate the effectiveness, efficiency, and response of a drug when targeting specific genes or when those genes are not targeted.
Such valuable information provides value in drug development, studies and discovery. It also strengthens the therapeutic approach against a disease.
Developing gene therapy:
Gene knockout recently has given more strength to the field of gene therapy. New therapeutic approaches and gene therapy treatments can be developed against particular or many gene defects.
Various CRISPR-cas9 therapies mediated by KO are now under clinical trials and ready to rule the world. Other therapies are also under research.
Agriculture and livestock improvement:
Not only in disease studies and human genetics, but gene knockout techniques also have great value in the agriculture industry and livestock improvement. Through knockout studies, researchers can understand the function of an economically important trait and thereby can enhance it.
Desirable and economically important traits like disease-resistant, yield improvement, nutrient values in plants and growth, development and milking capacity in animals can be improved.
Limitations of Gene Knockout:
Gene knockout is a highly useful and precise tool in the field of genetic engineering. However, like other techniques, it also has certain important limitations. Here are some limitations of gene knockout (KO):
Off-targeting effects:
Any gene knockout experiment is designed to specifically inactivate a target gene, however, oftentimes, it can suppress or affect other gene’s activity, unintentionally. Or in other cases, it inactivates entirely a different gene. These two scenarios are known as an off-targeting effect.
Research shows substantial off-targeting gene knockout effects, which may directly or indirectly impact the function of other gene(s) or regulatory elements.
Gene compensatory effects:
Compensatory mechanisms may exist for several genes that compensate or fill the function of an inactivated gene. This particularly occurs during the developmental process in which KO triggers the developmental compensatory mechanism.
Resultantly, even after successful gene knockout, the function will remain intact. Such events pose challenges to understanding the full potential effect of a gene and its governing traits.
Compensatory action:
In some cases, redundant genes can compensate for the impact of a target gene. Therefore, simply knocking out a single gene may not result in a loss of function. Other genes, known as redundant genes, can step in and continue to produce the expected traits. These compensatory effects of genes can restrict the effectiveness of gene knockout.
Gene knockout for essential genes:
There are a set of genes in organisms that are essential for their survival. Alteration in such genes can cause lethal effects or genetic abnormalities. Scientists can’t manipulate and study these genes by gene knockout.
Tissue specificity:
Gene knockout has less tissue specificity as it can impact a broad range of cells and tissues. So while targeting a specific gene for specific tissues, adequate results can not be obtained.
Wrapping up:
Gene knockout is an important technique in genetic engineering that helps scientists learn about the function of specific genes and how they work. It allows us to understand the mechanisms behind gene functions. Although there are some limitations like unintended effects, lack of specificity in tissues, and compensatory effects, scientists are actively working to overcome them.
Gene knockout is used in various fields such as agriculture, disease research, drug development, and other studies. Feel free to share this article and bookmark our page. Let us know your thoughts on this topic.
Sources:
- Scientists can analyse gene function by deleting the gene sequence. Nature; Essential of genetics unit 4.4 (link).
- Hall B, Limaye A, Kulkarni AB. Overview: generation of gene knockout mice. Curr Protoc Cell Biol. 2009; Chapter 19:Unit–19.12.17.