“Learn the basic concept of gene expression and epigenetics. Explore what epigenetics is and how it works. How different factors interact with our DNA and alter our epigenetic profile.”
Have you ever wondered why monozygotic twins with similar DNA, may look similar but act differently or have different behavior and even traits? Why do some traits like eye color, hair color, height and skin color, run in the family but have different expressions?
Why similar genes may produce variations in traits. ‘Genetics’ isn’t only responsible for all these events, certainly. So what exactly happens here? Here comes the role of ‘epigenetics.’
Epigenetics is a phenomenon of alterations in gene expression level and not gene sequence level. Meaning, a gene may produce its effect only when and where it is required. That’s directly involved in shaping our health, development and producing various traits as per the requirement.
Still looks confusing! Let’s dive into the world of ‘epigenetics.’ I am planning to prepare an entire series around this topic to explain different fundamentals and mechanisms. Starting with the very first article of this series– Epigenetics 101.
I actually don’t know how to start this article, trust me! Because there is a ton of literature and material available on the topic “epigenetics.” The idea behind writing this article is to give a comprehensive and simple explanation that is more useful to students!
Hope you like it! Explore what epigenetics is, and how it occurs along with some interesting facts and brief history, in this article.
Related article: Genetics vs Epigenetics: From Gene Alterations to Gene Expression.
Key Topics:
What is Epigenetics?
The term ‘epigenetic’ is made up of Epi + Genetics. The prefix ‘Epi’ was derived from the Greek word which means ‘around’ or ‘outside.’ This literally means, ‘outside or around genetics.’ In ~1942, Conrad Weddington coined the term ‘epigenetics.’
Weddington was a British biologist. He explained how genetics interact with the environment and produce different traits. In 1958, David Nanney officially used the term ‘epigenetics’ in his paper.
Between both these crucial findings, in 1948, Rollin Hotchkiss identified modified cytosine but the official paper explaining any chemical base modification viz DNA methylation was published in 1960. Afterward, the sub-discipline came to light. In 1970, histone modification was reported.
The epigenetic era actually began then after, slowly and gradually. And the picture becomes even more clear after the evolution of modern genetics– sequencing and NGS.
The entire field of genetics explains how a protein encodes from a gene. And everything is running around it, starting from replication to the final translation product. But epigenetics rather focus on ‘when’ & ‘where’ to form a protein and in ‘which amount.’
Meaning, it largely regulates ‘how much’, ‘when’ and ‘where’ a gene expresses or does its job! And that my friends, can directly impact a person’s health. To understand it more precisely, first understand some basic genetic concepts.
All our DNA is located on chromosomes. The process– from a singular DNA unit to a compact chromosomal structure is known as DNA packaging. Specialized DNA-binding proteins interact with the DNA and wrap them into a chromosome.
However, DNA may loosen or open up to allow various activities like replication, transcription or translation. Unpacking allows replication, transcription and translation and makes a gene ‘ON’ for producing a protein. Meaning, a gene expresses while Packing disallows any or all three activities and makes a gene ‘OFF.’
In addition, the entire process is also modulated by timely chemical modifications like methylation, histone modification, chromatin remodeling, etc. That’s how a gene’s activity can be regulated by a cell’s mechanism.
However, the packing and unpacking of DNA is a continuous process but still varies among cells or tissue.
Such modifications are broadly covered in epigenetic changes or modifications. More in-depth explanations suggest that extrinsic factors like environment, diet, mental health, stress and exposure to toxins, may directly not affect a gene at a sequence level but modify either protein-DNA interaction, DNA-binding factors or other chemical modifications in DNA and consequently alter gene expression.
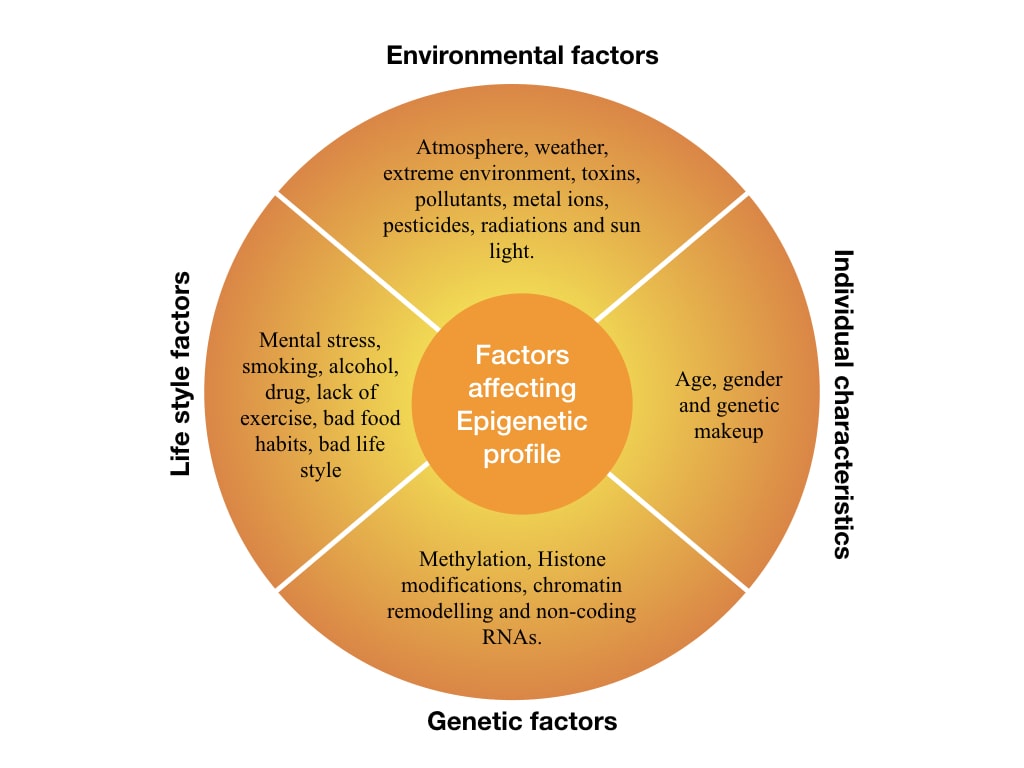
One study published in a Springer Journal suggests that exposure to pesticides can modify histone-DNA interaction while another study published by Oxford University Press suggests that stress or toxin exposure can directly affect DNA methylation patterns.
In both cases, pesticides or toxins alter a cell–, tissue– or organ–specific gene expression thereby the epigenetic profile of a person. Here is a list of factors that can impact epigenetics.
| Environmental factors | Genetic factors | Individual or lifestyle |
| Environmental pollutants | Methylation | Age |
| Environmental toxins | Histone modifications | Gender |
| Chemical exposure | Chromatin remodeling | Genetic makeup |
| Exposure to UV. X-ray and other harmful rays | Non-coding RNA action | Stress |
| Extreme temperature | Lack of exercise | |
| Heat or cold | Bad eating habits | |
| Pesticides | Smoking | |
| Metal ions | Tobacco consumption | |
| Ecosystem | Drugs | |
| Weather | Mental health | |
| Radiation | Work-life | |
| Pollution | Bad habits |
Besides these, there are many lifestyle and metabolic factors that can affect a person’s epigenetic profile. Conclusively, epigenetics is a complex and fundamental process governed by varieties of expressions like —personal habits, and extrinsic and intrinsic factors.
But when do genes need to turn on or off? Let’s find out.
All our body cells (except germ cells) have the same set of genes but not the same set of ‘active’ genes. Because not all the genes need to be active all the time and in all cells. For example, a gene set that is needed in brain cells and neuron development isn’t necessarily in live cells, and vice versa.
The ASPM gene produces new nerve cells in the brain while ADH1B is an alcohol dehydrogenase family gene that is required in the liver to metabolize alcohol. So it’s evident that the ASPM gene is highly active in brain cells while ADH1B is highly active in liver cells.
At the same time, ADH1B and ASPM are not readily required by brain cells and liver cells, respectively. So genes only activate and express when needed. In addition, genes may turn on or off by the influence of environmental factors.
Let’s take another example.
Skin, hair, and eye colors vary widely among people due to the production of melanin, a protein synthesized by specialized skin cells called melanocytes. The amount of melanin produced is determined by the expression of the melanin-coding gene, which leads to darker or lighter skin tones depending on the level of expression.
Melanin is important for us as it offers protection against harmful UV rays and skin cancer. Therefore, individuals living in warmer regions with more sun exposure tend to have higher levels of melanin and darker skin, while those living in colder regions have lower levels of melanin and fairer skin.
Interestingly, even though the sequence of the melanin-coding gene may be similar among all humans, its expression is influenced by various factors such as environmental conditions, which modify this fundamental trait. This highlights the importance of epigenetic adjustments and modifications in shaping human characteristics.
Read more: What is a Gene?- Definition, Structure and Function.
How do epigenetic alterations occur?
Methylation, histone modifications, chromatin remodeling and non-coding RNA processing are a few among many known epigenetic modifications. Let us take a simple mechanism that explains how epigenetic modifications occur– DNA methylation.
We will not go more in-depth, but rather we will focus on the concept. Ok, so a gene is a long DNA chain that can encode a protein. It is made up of introns, exons and regulatory sequences like a promoter, enhancers and regulators, etc.
Enzymes recognize such sequences and perform gene expression. At the chemical level, the protein or enzyme involved in the transcription (which is collectively a process of gene expression) identifies the chemical structure of bases, one by one, recognizes it, identifies it, processes it, settles on it and performs its action.
But what if a base’s chemical structure slightly changes, for example– if a chemical group binds with a cytosine base, The protein or transcription factor can not recognize the sequence, and consequently can’t execute its function.
Here, the gene fails to process and can not form a protein. A methyl group, when added to any of the bases but popularly high CG-rich regions, does the same job, and fails the process of transcription– down-regulates the gene expression.
Similarly, by any means, if a methyl group is detached from the DNA, it re-activates a gene, allowing the enzyme to work and perform transcription. This upregulates a gene expression. This can be said as epigenetic regulation mediated by chemical modifications.
Secondary structural modifications can also contribute to epigenetic regulation. For example, a simple histone modification. Again we will not go in-depth, just give a simple overview.
Histones are DNA-binding proteins that interact with the DNA, form a tight nucleosome structure and construct a chromosome. A tight wrapping of histones and DNA does not permit transcriptional factors to interact with the DNA, consequently, turns off gene expression.
Similarly, when histones loosen the DNA or are removed, it gives direct access to proteins and transcriptional factors to perform gene expression. This turns on gene expression.
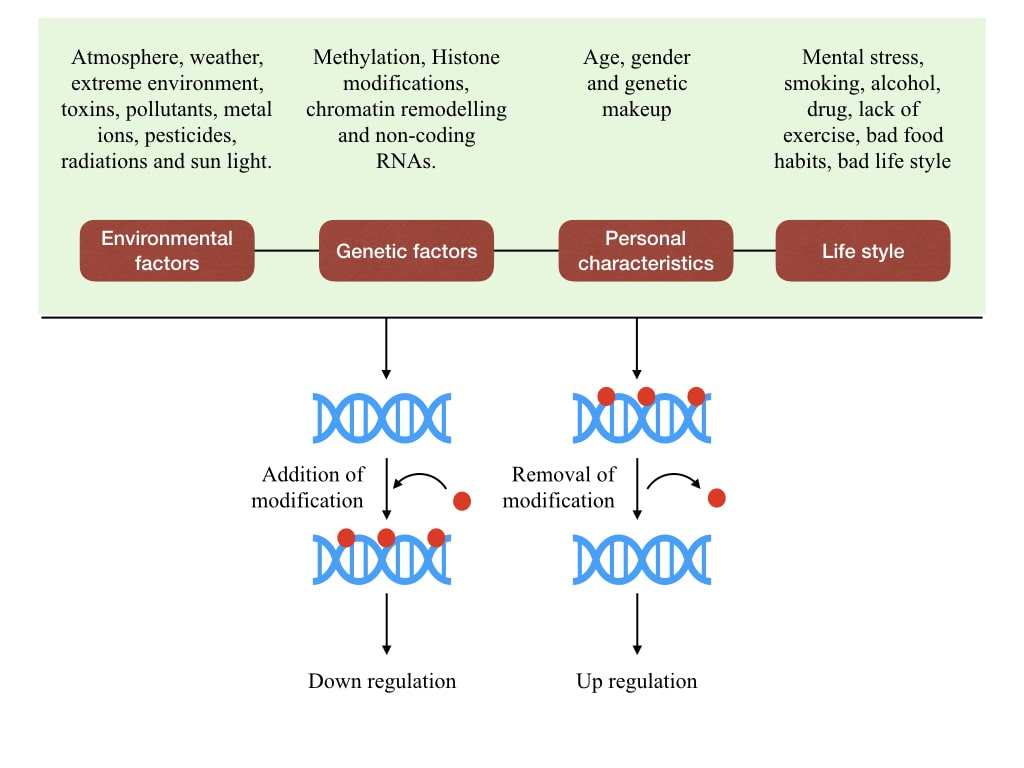
That’s how gene expression up– and downregulates. As aforesaid, there are many chemical and secondary structural modifications that occur to regulate the epigenetic process. We will discuss each one, in each separate article of this series.
Till now two things are, I guess clear, what exactly gene on and off is and how it occurs. You may wonder, are those changes heritable, like– genes and DNA? Or reversible? Unlike– structural genetic changes? Let’s find out.
Properties of Epigenetic alterations:
Heritability:
Parents pass down not only genes but also related alterations to their offspring, such as a mutation in a beta-globin gene. Similarly, epigenetic changes are also inherited as long as they occur in the germ cells and are ‘imprinted’ in the fetus.
But it is also crucial to note that these modifications can be influenced by the environment, genetics and lifestyle of a person. And may change throughout their lifetime.
Reversible:
Another crucial property of epigenetic modifications is ‘reversibility.’ It can be reversed. Unlike the sequence-level modifications which once fixed can’t be reverted, epigenetic changes can be re-programmed or improved.
Healthy lifestyle and environment and epigenetic therapies are a couple of options to repair or reverse epigenetic modifications.
Influenced by environment:
Epigenetic modifications are so sensitive and influenced by environmental factors and personal habits, as aforementioned. Exposure to toxins, pollutants, extreme conditions, food, diet, habits, stress and mental conditions all these factors can affect an individual’s epigenetic profile.
Dynamic:
Epigenetic modifications are dynamic. It can be inherited but can be reverted too. So throughout a person’s life, it constantly changes in response to external factors or stimuli.
Provides variability:
Yet another crucial property of epigenetic modifications is its ability to alter or improve phenotypes. Such changes give rise to phenotypic variability for a single trait.
Take the example of skin colors or shades, eye colors or shades, height, weight, and even mental strength; all these phenotypes can be produced from a gene or group of similar genes but the different versions of the trait are produced by epigenetic modifications. I already explained how it occurs.
Note that sometimes, sequence-level alterations may also contribute to phenotypic varieties.
Local:
Epigenetic modifications are local. Each cell or tissue has its epigenetic profile viz. collection of highly, moderately and inactive genes. Different tissues can exhibit different epigenetic profiles for their functions.
Sensitive:
Epigenetic modifications are so sensitive. A small change in lifestyle or surroundings can exhibit up or down-regulation of genes. The highly sensitive nature of epigenetic changes produces many diseases like diabetes or cancer.
Wrapping up:
Conclusively, epigenetic modifications are crucial for a person’s health. Changes in gene expression can cause serious health conditions like cancer. P53 is one candidate gene whose expression, when altered, causes various types of cancers.
Interestingly, epigenetic changes can be revered and thus a person can actually go for epigenetic therapy just like rehabilitation, and can improve their genetic representation by adopting a good lifestyle.
I hope you like this article. This series will continue. In upcoming articles, we will discuss methylation, histone modifications and many more things.
Sources:
Deans, Carrie, and Keith A Maggert. “What do you mean, “epigenetic“?.” Genetics vol. 199,4 (2015): 887-96.


Good article
Thank you for appriciation