A complete history of DNA sequencing and key milestones are discussed in this article.
Sequencing has been a significantly important technique in genetic research for years. However, Sanger sequencing has exceptional value compared to other techniques.
Other sequencing platforms like Maxam-Gilbert, pyrosequencing, SOLiD sequencing, and high throughput sequencing platforms evolved timely but failed to compete and rise.
In this article, we will revisit the history of various sequencing platforms.
Stay tuned.
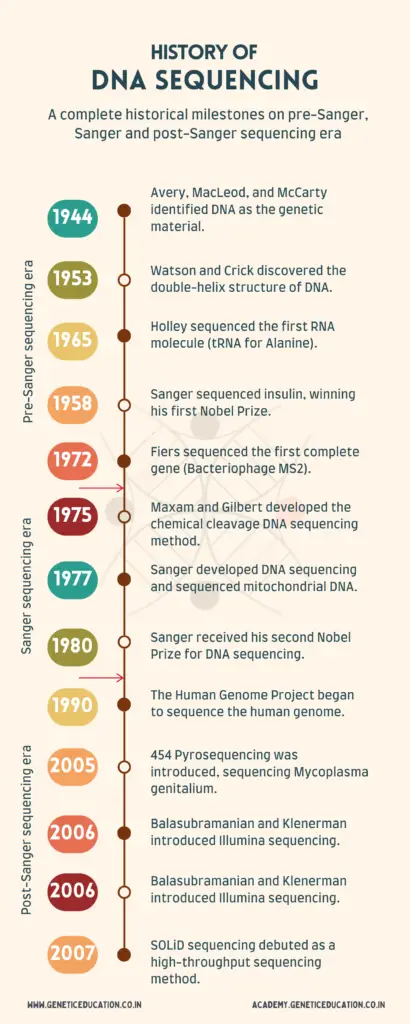
We will divide this article into three parts: the pre-Sanger sequencing era, the Sanger sequencing era, and the post-Sanger sequencing era. Keep in mind that we will only discuss some major milestones in the sequencing industry.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy to understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Key Topics:
Pre-Sanger Sequencing Era:
Before starting to sequence, it is important to know the history of DNA as genetic material.
In 1944, Oswald Avery, Colin MacLeod, and Maclyn McCarty experimented on Streptococcus pneumoniae bacteria to determine what is responsible for transformation.
Their experiment validated that DNA is the genetic material responsible for transformation.
In 1953, Watson and Crick identified the DNA double helix and also postulated the chemical structure of the DNA.
The first sequencing milestone was achieved in 1965 when scientist Robert W. Holley sequenced the first RNA molecule (tRNA) for Alanine from Saccharomyces cerevisiae.
This was the first time any biological molecule was sequenced.
In 1972 another scientist named Walter Fiers achieved another milestone by sequencing the DNA of a complete Bacteriophage MS2 gene.
Sanger Sequencing Era:
Now, between the 70s and 80s scientists started exploring how to sequence DNA and two important discoveries floated out as innovative and useful approaches.
We are talking about the Sanger sequencing era. And milestones achieved during this period.
Sanger sequencing was developed by Frederick Sanger in 1977, development of this method revolutionized the field of molecular biology. This technique was the first by which scientists were able to determine the sequence of the nucleotides in the genome precisely.
Here are the glims of Sanger’s approach.
Sanger was initially working with the sequencing of proteins. In the area of protein sequencing, he determined the sequence of Insulin for which he was given the Nobel Prize in 1958.
Later on, Sanger started working on the sequencing of RNA. As RNA is single-stranded it was much easier to sequence. These two sequencing efforts later established a strong foundation for DNA sequencing.
He started working with the basic principle of DNA replication where DNA polymerase adds the dNTPs in a newly synthesized strand. For sequencing, he used Template DNA, Primers, DNA polymerase, dNTPs, and ddNTPs.
He used the idea of the basic replication process. Here, the DNA polymerase uses nucleotides and adds to the growing DNA strand by utilizing the free 3’ OH end of the RNA primer.
Sanger followed the exact scheme and achieved the sequencing milestone with a small modification. He used ddTNPs as well to generate fragments.
To know more about the complete mechanism you can read our previous article on Manual vs Automated Sanger sequencing.
Later on, the human mitochondrial DNA and bacteriophage λ were sequenced by Sanger and his colleagues.
Sanger was awarded the second Nobel Prize for the present achievement in 1980.
Advancements in Sanger sequencing like the use of fluoro-labeled ddNTPs, capillary gel electrophoresis, use of automated and software-based data analyses outranked all their competitors over time.
Resultantly, it became the gold-standard method and used to sequence the human genome during the HGP.
In 1990 Human Genome Project was initiated to determine the nucleotide sequence of the complete human genome with the help of Sanger sequencing.
In 1975 Allan Maxam and Walter Gilbert used the chemical cleavage technique for DNA sequencing.
Although the Maxam and Gilbert technique used the same polyacrylamide gels to resolve DNA fragments, their approach differed dramatically.
Instead of using DNA polymerase to generate fragments, radiolabeled DNA is treated with chemicals that break the chain at specific bases; after running on a polyacrylamide gel, the length of cleaved fragments (and thus the position of specific nucleotides) can be determined and the sequence inferred.
These two approaches were the first that were extensively accepted and hence might be considered the true beginning of ‘first-generation’ DNA sequencing.
The first generation of sequencing was able to read the nucleotide sequence less than 1 kilobase in size.
Related article: First, Second and Third Generation of Sequencing.
Post-Sanger Sequencing Era:
The high throughput sequencing era began with the introduction of 454 pyrosequencing in 2005. The present method is based on the luminescent that detects the pyrophosphate released during the synthesis process.
A two-enzyme process in which ATP sulfurylase converts pyrophosphate into ATP, which is then used as the substrate for luciferase, resulting in light proportional to the amount of pyrophosphate.
This method was utilized to determine sequence by monitoring pyrophosphate detection while each nucleotide is washed through the device.
This was the first high-throughput sequencing technique that was capable of sequencing a few thousand bases in a single run.
Using this technique the genome of Mycoplasma genitalium was successfully sequenced in 2005. Pyrosequencing was subsequently licensed to 454 Life Sciences, a biotechnology business formed by Jonathan Rothburg.
Later on in 2006 Shankar Balasubramanian and David Klenerman at Cambridge University introduced another sequencing technology named Illumina or Solexa sequencing.
In this technique, DNA is fragmented, and adapters are ligated onto the fragments then Fragments are connected to a flow cell and amplified using bridge PCR.
Sequencing-by-synthesis is carried out with fluorescently labeled reversible terminators. A high-definition camera records photos of fluorescent signals, which are then used to decode the sequence.
These techniques were considered the second generation of sequencing or next generation sequencing, along with these two SOLiD Sequencing approaches were also introduced in 2007,
SOLiD sequencing (Sequencing by Oligonucleotide Ligation and Detection) was developed by Applied Biosystems as a high-throughput sequencing platform at that time.
The present technique also works on the principle of sequencing by synthesis (which we explained in detail in our previous article).
Here, fluorescently labeled oligonucleotide probes are ligated to complementary DNA fragments, and upon ligation these probes generate fluorescent signals. These signals are then decoded to determine the sequence.
Although SOLiD sequencing failed to penetrate the industry, it gained popularity and opened doors for high-throughput and ultra-high-throughput sequencing platforms.
Wrapping up:
These are a few important milestones in the sequencing industry. However, keep in mind that no platform can work if polymerase, ddNTPs, ligase, and other smaller discoveries were not made.
We will discuss that in some other article. This foundation will help us to understand how long it will take us to sequence a complete genome within a day.
I hope you enjoyed reading this article! Explore our sequencing courses to gain in-depth knowledge about sequencing techniques, practical applications, and data analysis.
Resources:
Heather, J. M., & Chain, B. (2016). The sequence of sequencers: The history of sequencing DNA. Genomics, 107(1), 1–8.