“A Mitochondrial DNA test has been utilized to study and trace the matrilineal line, but how do they do that? Learn the concept and procedure of mtDNA testing in this article.”
Mitochondria is a cell’s powerhouse. It’s a membrane-bounded organelle present in the cell cytoplasm. The presence of DNA makes it unique from other cell organelles. This DNA is different from a cell’s nuclear DNA, known as mitochondrial DNA.
Another interesting thing about the mitochondrial is that the DNA present here can only be inherited from the maternal side. Meaning, from the mother only, although, it is present in both— males and females.
Therefore, testing mtDNA can help understand and prepare the maternal lineage accurately. This means one can know that traits and genotypes are inherited from his or her mother.
mtDNA test is pivotal for forensic science and ancient DNA studies. But how do they test mtDNA? How does the test work and what is the procedure? Let me explain it in this article.
I’m Dr Tushar Chauhan and I’m a geneticist and expert in genetic testing techniques. I will explain the procedure of mtDNA testing from the expert’s point of view in an easy-to-understand manner.
Stay tuned.
Related article: How and Why is mtDNA Typing Used in Forensic Science?
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
Mitochondrial DNA Test Steps and Procedure
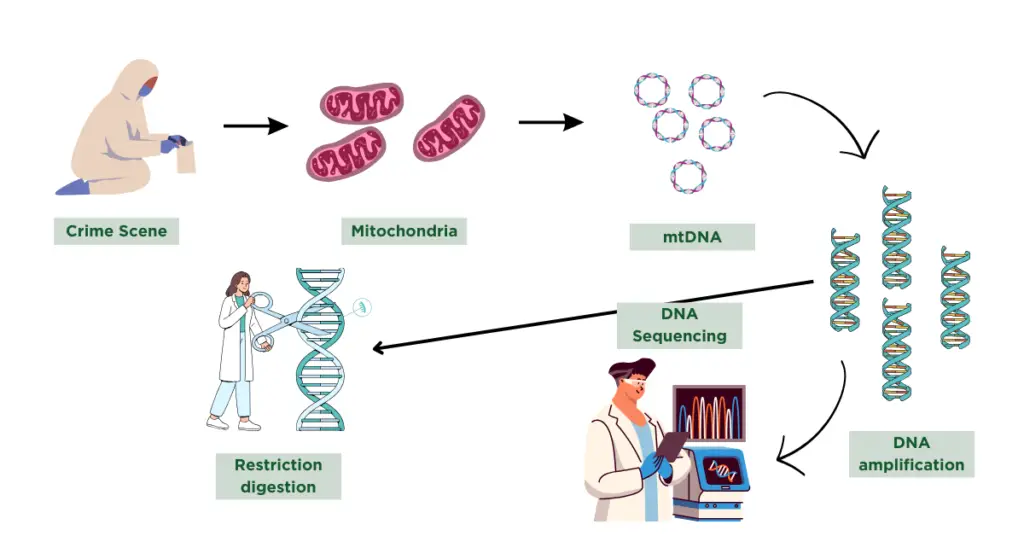
Sample collection, DNA extraction, DNA quantification and quality assessment, DNA amplification, fragment analysis, Results and interpretation are several common steps in which the entire mtDNA testing procedure can be divided.
I will explain each step comprehensively in this section.
Sample collection:
mtDNA can be isolated from a variety of samples including buccal swabs, hair, bone, teeth or even blood spots. Any nucleated cells can be employed for mitochondrial DNA extraction.
Various dedicated procedures and kits are available to isolate DNA from a variety of samples. However, conventional methods such as Proteinase K or phenol, and chloroform DNA extraction methods are employed as well.
Once adequate DNA has been obtained, it is sent for quality and quantity assessment. Note that the sample should be collected, stored and transported under adequate conditions. Refer to the sample handling guide for that.
Related articles:
- Proteinase K DNA extraction method.
- Phenol Chloroform DNA Extraction: Basics, Preparation of Chemicals and Protocol.
DNA quality and quantity assessment:
DNA quality and quantity assessment is a crucial pre-step before processing a sample for amplification and sequencing. A good quality DNA extract should have a decent purity and adequate quantity to use it.
Instruments like Nanodrop Lite, Qubit, Qiagen QiaXpert and Bioanalyzer can be used.
PCR amplification:
PCR amplification has been performed to enrich the DNA fragment being studied. The isolated mtDNA is not adequate to be directly used for DNA sequencing or restriction digestion.
Using a standardized PCR protocol, copies of HVR1, HVR2 and other coding regions have been prepared. To do so, the DNA is amplified using the sequence-specific primer sets.
After successful amplification, fragment libraries are generated to use it in DNA sequencing. The sample can be processed using either DNA sequencing, which is the most robust, sensitive and accurate method and RFLP which is the conventional way to do it. We will understand both the techniques one by one.
mtDNA analysis:
RFLP:
RFLP is Restriction Fragment Length Polymorphism. In this method, the amplified mtDNA fragments are digested using a specific type or a set of specific restriction endonucleases.
The sample being tested and the sample used as a reference both, are digestion using the same set of restriction enzymes. A restriction enzyme will digest the fragment if its specific recognition site is present.
Each REase will work in the same fashion and can produce fragment polymorphism in a gel. These fragments are compared with the reference sample for interpreting results. HpaI, BamHI, HaeII, MspI, AvaII, and HincII are common REases used in mtDNA testing.
DNA sequencing:
As aforementioned, DNA sequencing is the best, most trusted, accurate, sensitive and robust mtDNA analysis method. It can even identify a single nucleotide alteration from the sequence and provides us with in-depth data for maternal tracing.
Sanger sequencing has been employed for forensic, ancient and disease-related mtDNA tests for a long time. It’s a simple yet effective chemical-chain termination method in which once the polymerase encounters the ddNTP, it terminates the chain synthesis.
So after each nucleotide when a specific ddNTP is incorporated, the chain is terminated and the sequence can be read depending on the ddNTP incorporated. The present sequencing platform can identify common and new mtDNA polymorphisms present in various populations.
However, on the downside, Sanger sequencing can effectively read shorter sequences of a few thousand base pairs.
The recently evolved NGS (Next-Generation Sequencing) platforms are high throughput, ultra-sensitive, robust and accurate compared to the conventional Sanger sequencing approach.
They can detect common and novel mitochondrial mutations, length variations and indels present in the mitochondrial genome. Thus, NGS can be used in forensic and ancient DNA studies as well as mitochondrial disease research and screening.
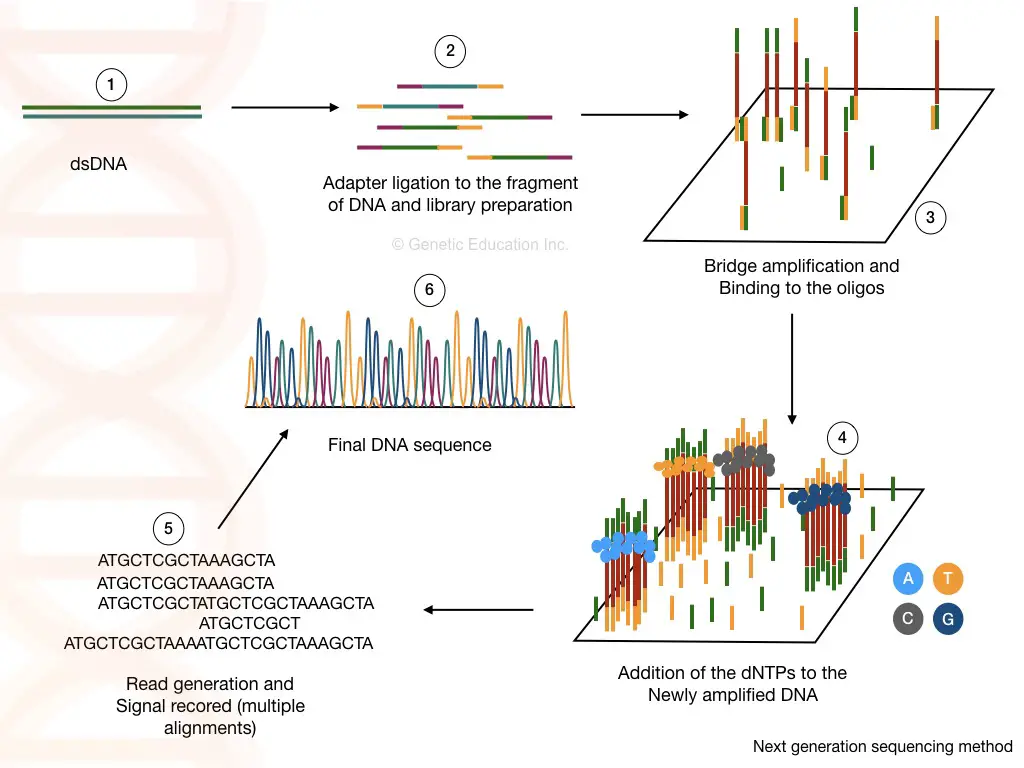
In addition, it can accurately study the D-loop DNA (hypervariable regions) and sample heteroplasmy precisely. Thus, NGS is widespread nowadays in mitochondrial DNA testing and research.
Two types of testing approaches are low-resolution and high-resolution sequencing. The low-resolution sequencing includes HVR1 and HVR2 while the high-resolution sequencing includes HVR1, HVR2 and coding regions.
High-resolution sequencing is often known as full or complete mtDNA sequencing as well which is ideally recommended to accurately trace the maternal lineage.
Massive parallel sequencing, Cycle sequencing, pyrosequencing, etc, are used for mtDNA sequencing.
| RFLP for mtDNA testing | Sequencing for mtDNA testing |
| Can study fragment length polymorphism. | Can study fragment length polymorphism. |
| Identify known mtDNA mutations. | Identify known and novel mtDNA mutations. |
| Can study length heteroplasmy. | Can study length heteroplasmy at the sequence level. |
| Manual, less sensitive and low throughput. | Robus, sensitive and high throughput. |
Results:
RFLP results can be studied using either PAGE or conventional agarose gel electrophoresis. However, it needs an expert’s opinion for results.
Conversely, the results of the sequencing can be studied using a computational program. Once the machine completes the sequencing reaction, the sequence data is transferred to the computer program.
The program helps decode the sequence information and variations. Heteroplasmy, length polymorphisms, SNPs and indels can be identified and reported.
Analysis and interpretation:
To analyze and interpret the mtDNA results is a complex and tedious process. No two mitochondrial genomes are completely similar in the world! Otherwise, everyone would be similar.
Mutations such as SNPs, indels and length variations make each mtDNA unique. All these variations have been studied for tracing the maternal lineage. To do so, first, the sample sequence is compared with the reference sequence.
Two commonly used mtDNA reference genome sequence databases are the revised Cambridge Reference Sequence (rCRS) and the Reconstructed Sapiens Reference Sequence (RSRS).
Based on the mutations present in the sample, its match with the reference databases, and using a complex computational analysis a matrilineal map of the sample has been prepared.
In the case of a forensic sample, the sample sequence is matched against the reference sequence to determine the possible close maternal relative. And for disease studies and research, the same database has been used to link the mutations with a particular type of disease condition.
As I said, interpreting the mtDNA test results is a complicated process, we will understand it comprehensively in another article. So this is the complete process of a typical mitochondrial DNA test whether it is for forensic, ancient DNA study, disease research and diagnosis purposes.
Related article: 8 Reasons Why Is Mitochondrial DNA Inherited Maternally?
Wrapping up:
mtDNA testing or typing is an important tool only when the nuclear DNA is not available or degraded. However, to understand one’s matrilineal history, it can be ordered. Techniques like next-generation sequencing revolutionized the testing field, though, the data provided don’t accurately tell us about the lineage history.
yDNA test and nuclear DNA test data are required to accurately understand a person’s ancestry history. Still, it is worth it, only if you would like to know about your mom, her mom, her grandma’s mom and so on.
I hope you like this article.
Sources:
de Benedictis, G et al. “Restriction fragment length polymorphism of human mitochondrial DNA in a sample population from Apulia (southern Italy).” Annals of human genetics vol. 53,4 (1989): 311-8. doi:10.1111/j.1469-1809.1989.tb01800.x.
Comparative analysis of mitochondrial genome by NGS from Illumina.