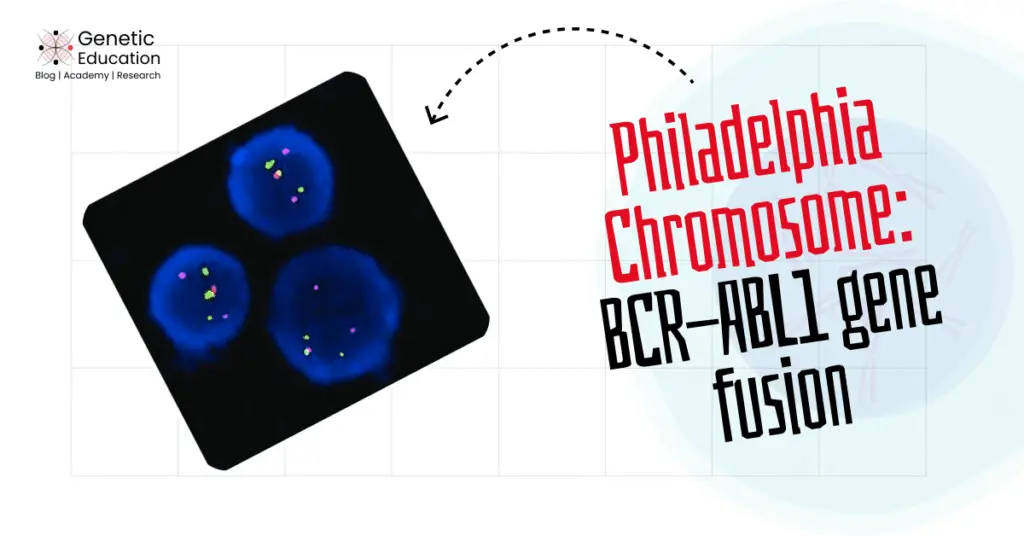
“The Philadelphia chromosome occurs due to the translocation between chromosomes 9 and 22, resulting in the fusion of the BCR-ABL1 genes. Explore this complete article on the genetics of the Philadelphia chromosome.”
Numerical and structural chromosomal abnormalities are involved in various types of cancer, deciphering how genetic factors cause cancer. Translocations are one such structural anomaly often reported in cancer.
The classic case of translocation is the Philadelphia chromosome, resulting from the gene fusion of BCR and ABL1 genes and leading to CML (chronic myeloid leukemia).
Learn the complete concept of the Philadelphia chromosome, how it occurs, its causes, and detection techniques. In addition, the molecular mechanism of gene resulting gene fusion is also explained here.
Stay tuned.
Key Topics:
What is the Philadelphia chromosome?
The Philadelphia chromosome is a product of chromosomal translocations involving chromosomes 9 and 22. It is a cancer-causing genetic aberration.
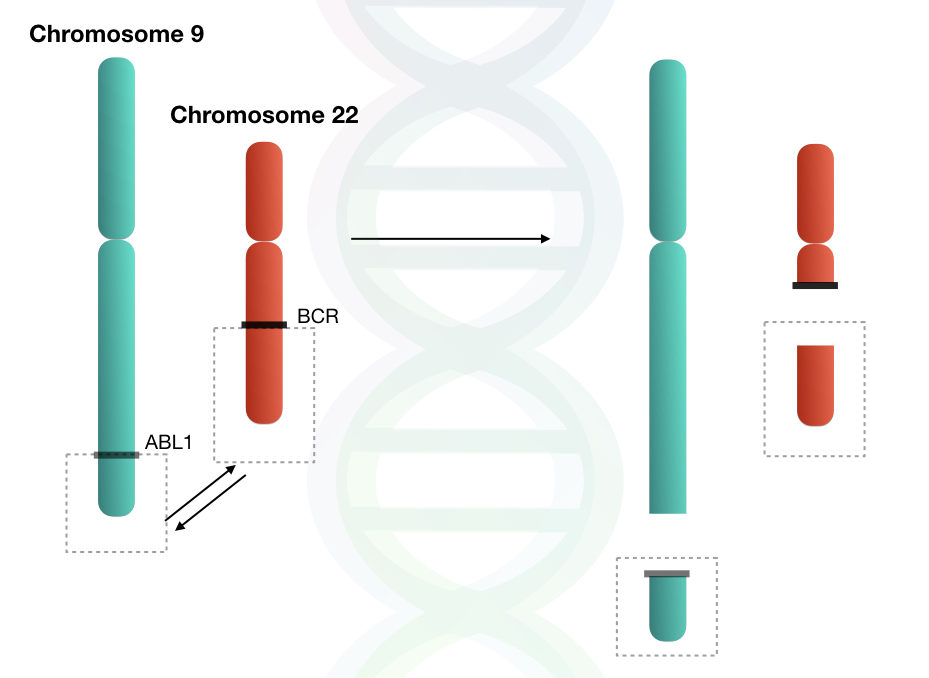
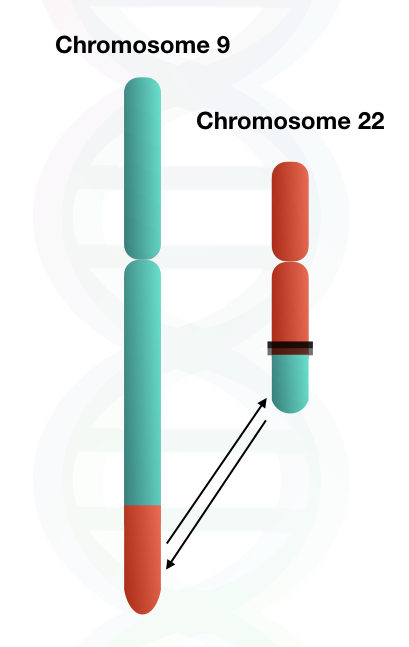
The reciprocal translocation between chromosomes 9 and 22 results in the shortened chromosome 22, which is referred to as the Philadelphia chromosome. This translocation involves the long arms (q arm) of both chromosomes.
The chromosome 22 is called the Philadelphia chromosome, which, when diagnosed by karyotyping, is seen as truncated or shortened under a microscope.
The reason for the truncated or shortened chromosome 22 is the translocation of the BCR gene. The translocation not only involves the BCR gene but also involves distal portions of chromosome 22 along with the BCR gene.
The BCR gene, along with distal portions either incorporated into chromosome 9 or lost during translocation. Therefore, the changes in the size of chromosome 9 are not noticeable by microscopy.
But in the case of chromosome 22, a large portion is being translocated, so it results in a significant decline in the size of chromosome 22.
Let’s not dig in further on why the size of the chromosome decreases, we will discuss that in a separate article in the future.
So, in short, this translocation results in the fusion of two important genes, the BCR gene of chromosome 22 and the ABL1 gene of chromosome 9. This major event is reported in chronic myeloid leukemia.
Historical events:
The studies of chromosomal aberrations began in the early 19th century when Karl Sax observed the first ever chromosomal aberration using X-rays.
In 1959, during cytogenetic studies of leukemia patients, David Hungerford and Peter Nowell discovered the Philadelphia Chromosome.
After several years of study, in 1973, Janet Rowley found that the Philadelphia chromosome is a result of reciprocal translocations, and he also identified the genes that are involved in this genetic alteration.
She used high-resolution Karyotyping to confirm this reciprocal translocation. Interestingly, before Janet Rowley’s confirmation, researchers believed that the Philadelphia chromosome was due to deletions.
Moreover, after a few years, it was identified that the ABL1 gene is a protooncogene that has tyrosine kinase activity.
Later on in 1990, researchers named Nora Heisterkamp and her colleagues published the paper that provided the evidence for the BCR-ABL1 gene fusion and its involvement in cancer (Chronic Myeloid Leukemia-CML).
What are the BCR and ABL1 genes?
BCR and ABL1 genes are also known as BreakPoint Cluster Region (BCR) and Abelson (ABL). These are the two important genes, in which BCR is located on chromosome 22 and ABL1 is located on chromosome 9.
Let’s discuss each gene one by one to understand its roles and functions better.
BCR (Breakpoint Cluster Region) gene:
The BCR gene is located on the long arm of chromosome 22, which has 130 kb gene size.
The BCR gene encodes for the BCR protein, which is important for intracellular signaling and cytoskeletal organization. BCR protein also acts as a regulatory and scaffold protein.
The BCR gene has the coiled-coil domain, which can enable dimerization and oligomerization. When the BCR gene fuses with the ABL1 gene by reciprocal translocation, this coiled-coil domain forces oligomerization.
That results in permanently switching on the tyrosine kinase of ABL1.
ABL1 (Abelson) gene:
The ABL1 gene is located on the long arm of chromosome 9, which has 230 kb gene size.
ABL1 is one of the important genes for cell cycle regulation, stress response, and DNA repair, as it encodes a non-receptor tyrosine kinase that is involved in multicellular processes.
In the ABL1 gene, there is one important domain named SH3. In the normal ABL gene, this domain can auto-inhibit tyrosine kinase activity. But when it is fused with BCR, SH3 loses its inhibitory activity, leading to autophosphorylation of tyrosine residues.
Phosphorylated tyrosine residues lead to activation of oncogenic signaling pathways, resulting in cancer, such as Chronic Myeloid Leukemia (CML).
Molecular mechanism of BCR-ABL1 gene fusion:
Chromosomal translocations are the hallmark of certain cancers, such as CML, that occur due to reciprocal translocation resulting in the gene fusion of two important genes, BCR and ABL1.
The translocation between chromosome 22 and chromosome 9 is also known as t(9;22)(q34;q11), which involves the long arms of both chromosomes.
Note: What is t(9;22)(q34;q11)?
It is a cytological nomenclature to denote the translocation.
- T- Translocation
- (9;22)- between chromosomes 9 and 22
- (q34;q11)- between the q arm band 34 of chromosome 9 and the q arm band 11 of chromosome 22
This reciprocal translocation gives rise to a new fusion gene called the BCR-ABL1 fusion gene. So it is necessary to understand how this gene fusion occurs between two different chromosomes.
One of the most frequently observed gene fusion mechanisms is Non-Homologous End Joining (NHEJ).
Breaks occur in the double-stranded DNA within these two chromosomes, and these breaks are incorrectly joined by NHEJ, resulting in a new fusion gene, the BCR-ABL1 gene.
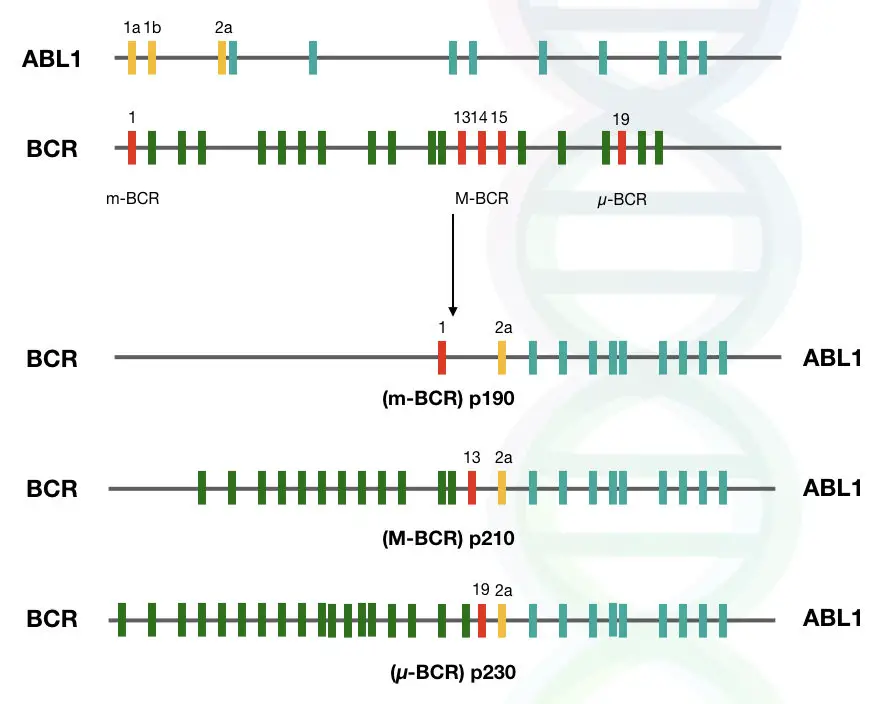
Protein variants and breakpoints:
As we discussed in the previous section, gene fusion is initiated by double-stranded breaks and then incorrect rejoining. These breaks can occur at different parts of the genes.
So it is important to know which are the common breakpoints in these two genes, because different breakpoints will give rise to different transcript variants, and eventually, they will be transcribed into different protein variants.
In the case of the ABL1 gene, breaks occur mostly at intron regions, such as near intron 1 and intron 2.
On the other hand, breaks can occur at three different regions in the case of the BCR gene. According to how often breaks occur in a particular region, breakpoint regions are classified in three categories that are listed below:
- Major breakpoint cluster region (M-bcr) – The Majority of the breakpoints occur in this region, which includes introns 12 to 16.
- Minor breakpoint cluster region (m-bcr) – In some rare cases, breaks occur at this region; however not commonly seen, and it includes exon 1 of the gene.
- Micro breakpoint cluster region (μ-bcr) – In this region of the gene, breaks do not occur.
These different breakpoints, when rejoined incorrectly by NHEJ, will give rise to different protein variants, such as p210, p230, and p190.
How is the Philadelphia chromosome linked with cancer?
As we discussed till now, the Philadelphia chromosome results from a reciprocal translocation, which gives rise to the fusion gene BCR-ABL1 on chromosome 22.
This fusion gene is not seen in normal individuals, as it is a hallmark structural genetic aberration leading to several blood cancers. The Philadelphia chromosome was the first genetic abnormality that was found to be linked with cancer.
The fusion gene resulting from translocation encodes for the chimeric protein, which has two portions, one from the BCR gene and the other from the ABL1 gene.
The BCR gene portion largely encodes for the dimerization as well as autoactivation of tyrosine residues by its coiled-coil domain.
Moreover, the ABL1 gene portion introduces the tyrosine kinase, which regulates the activity of tyrosine residues in normal cells.
In the fusion gene, it loses its activity, but due to autophosphorylation, tyrosine kinase activity remains continuously active. So, in simple words, the BCR gene portion removes ABL1 gene’s autoinhibition activity, which will lead to the activation of cell signaling pathways.
This will promote cellular proliferation, inflammation, rapid cell cycle progression, and the inhibition of apoptosis. These factors directly promote cancer.
The ABL1 gene is also involved in DNA damage response and repair mechanisms, so the BCR-ABL1 fusion gene will disrupt this process, which can lead to genetic instability.
Detection Techniques
Over the years, the Philadelphia chromosome has been detected by cytogenetics and molecular techniques such as Karyotyping, PCR, FISH, and Sequencing. Each method has different working principles, advantages, and disadvantages, so let’s discuss each one by one.
Karyotyping:
Karyotyping remains a gold standard technique for chromosomal studies. It’s a cytogenetic technique scientists have been using for Philadelphia chromosome studies for a long time.
It can spot the translocation between chromosomes 22 and 9 as the shorter q of chromosome 22 and the longer q arm of chromosome 9. However, expertise in cytogenetics and experience working with cancer karyotyping are required to study and interpret such complex rearrangements.
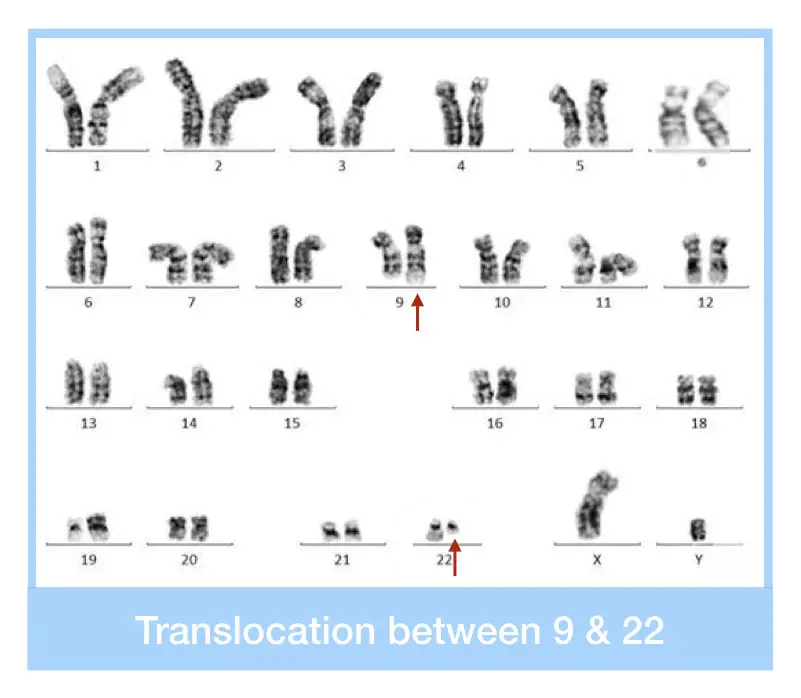
Due to the availability of the FISH, karyotyping nowadays has been used as a primary screening technique as it is the least accurate, time-consuming and tedious. In addition, an extensive cell culture setup and procedure are also needed.
FISH:
FISH (Fluorescence In Situ Hybridization) is a high-resolution karyotyping technique. It enables scientists to investigate structural chromosomal alterations effectively. And considered a gold-standard technique for cancer cytogenetic studies.
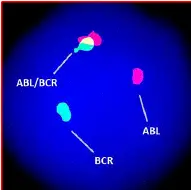
FISH— the probe-based hybridization approach uses fluorescence technology for detecting chromosomal alterations at a sequence level. For instance, two different colored probes are used for the BCR-ABL1 gene fusion.
The mixed fluorescence signals indicate the translocation. The present technique is more accurate, sensitive and reliable. In addition, it doesn’t need any culture or harvesting steps.
Ready-to-use cancer probes are now available, which save time and increase the efficiency of testing. The FISH test is highly recommended to study complex chromosomal alterations like the Philadelphia chromosome.
Note that FISH is also used to monitor treatment response in CML patients to know if the treatment is working or not!
PCR:
PCR (Polymerase Chain Reaction) is employed to validate the BCR-ABL1 gene fusion. However, it can’t directly provide chromosome-level information, instead, it provides quantitative data regarding the gene fusion activity.
Primers that are specific to the BCR-ABL1 flanking region can be designed to amplify the BCR-ABL1 fusion gene. PCR will only show the amplification if the BCR-ABL1 fusion gene is present; otherwise, no amplification will be seen.
The advanced quantitative RT-PCR provides quantitative BCR-ABL1 gene fusion data at a transcript or mRNA level and helps determine cancer progression and stages.
PCR is a fast, accurate and cheaper option for the Philadelphia study. PCR is capable enough to study sequence-level alterations. Still, FISH is the primary and gold-standard technique for studying translocations and gene fusion.
DNA sequencing:
DNA sequencing isn’t the first choice for BCR-ABL1 gene fusion detection in diagnosis, but it can be employed for research. Several research papers used the sequencing approach to study the gene fusion activity.
The amplified fusion product has been sequenced to identify the breakpoint and to study or spot new alterations present within the fusion gene.
Wrapping up:
Advanced cancer genetic testing techniques, available in recent times, are capable enough for identifying and diagnosing complex conditions like the Philadelphia chromosome.
The present condition is non-inherited, occurring randomly without any evident cause. The BCR-ABL1 gene fusion induces oncogene activities leading to cancer progression.
I hope you like this article. Do share it and subscribe to Genetic Education.
This article was updated by Vijay Odedara, who holds a bachelor’s and master’s degree in genetics. He brings extensive experience in the fields of genetics and cancer cytogenetics.
References:
Ayatollahi H, Keramati MR, Shirdel A, Kooshyar MM, Raiszadeh M, Shakeri S, Sadeghian MH. BCR-ABL fusion genes and laboratory findings in patients with chronic myeloid leukemia in northeast Iran. Caspian J Intern Med. 2018 Winter;9(1):65-70. doi: 10.22088/cjim.9.1.65. PMID: 29387322; PMCID: PMC5771363.
Hooberman AL, Westbrook CA. Molecular methods to detect the Philadelphia chromosome. Clin Lab Med. 1990 Dec;10(4):839-55. PMID: 2272177.
Haider MZ, Anwer F. Genetics, Philadelphia Chromosome. [Updated 2023 Jul 17]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK560689/.
Kang, ZJ., Liu, YF., Xu, LZ. et al. The Philadelphia chromosome in leukemogenesis. Chin J Cancer 35, 48 (2016). https://doi.org/10.1186/s40880-016-0108-0.