“Resequencing is a process to re-analyze a particular genomic region, gene or variant for validation. Learn about the re-sequencing process and why it’s needed.”
Sequencing is a complex process, particularly the whole genome sequencing. In WGS, we have to sequence nearly 3.2 billion base pairs, when talking about the human genome. That’s certainly a time-consuming, costly and tedious process.
Targeted or gene sequencing aims to read a gene or fragment from the genome, while a WGS’ aims to sequence an organism’s entire genome. Thus, WGS provides comprehensive, extensive and more sight from the genome.
However, the more the data generated- the sequences read, the more the chances of error. WGS, hence, is an error-prone technique. Various unread, gap, unclear and complex regions remain under doubt and require more clarity.
To study and understand such regions, resequencing is usually needed. In this article, I will explain what resequencing is and why and when it is required.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is resequencing
Resequencing is a process to reanalyze a particular region, gene, variant or group of variants for validation. A subset of genes or a group of disease-associated variants are analyzed using either Sanger or the next-generation resequencing approach.
I reviewed much literature on the Internet that mainly defines resequencing as a process to align the sequencing reads with the reference genome. But in clinical diagnosis, we used ‘resequencing’ when we actually needed to reanalyze a sequence either due to lack of clarity or to validate the finding.
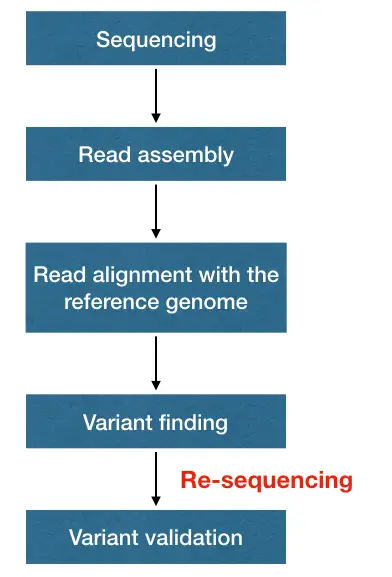
Now first let’s understand why it’s required.
Why Resequencing is required?
Let’s take an example.
Targeted resequencing
Case 1: let’s say scientists are studying the role of a novel gene associated with ‘some’ types of cancer. They observed a heterozygous base calling at two different locations in a gene during the targeted sequencing approach.
The gene is a bit complex in structure, considering the sequencing limitations of the Sanger sequencing technique, before reporting the variation it must be validated. For instance, one particular variation is an SNP after a long repetitive sequence block and another variation is a 3 base pair deletion of TTT nucleotides.
Overall, both variants are difficult to report without validation. So to get more clarity scientists have to resequence both the regions individually or in a single run, using either the same Sanger technique or NGS.
Only after resequencing validation the gene variations can be said as “associated with the disease or cancer.”
Thus targeted resequencing is employed to study gene variants associated with a disease, study the exonic regions or entire exon, study the whole set of coding regions associated with a disease and validate novel gene variants and heterozygous conditions.
Whole genome resequencing
Case 2: Let’s say scientists are doing whole-genome sequencing to understand the role of various genes or genomic regions associated with cancer.
Resequencing here is used to study SNPs, insertions & deletions and complex genomic regions. It’s also used to fill the gaps during WGS. However, to use resequencing for the WGS report is a bit tricky.
For instance, we can find many gaps, unknown variations (SNPs and indels) and complex and incorrectly sequenced regions from the whole genome. Now, it’s difficult to reanalyze (meaning resequence) the entire pool of “uncertainty.”
The reason is that it is a tedious, time-consuming and costly process. So what we do in a lab is understand the requirements for WGS.
For instance, if the sample is collected for colon cancer study. We first collect information regarding the genes and genomic regions associated with colon cancer and accordingly collect the information. Afterward, we used to do resequencing for those sets of genes and genomic regions.
NGS platform as well as Sanger sequencing, can be used for whole-genome resequencing. However, NGS can give us re-sequencing information regarding many genes or genomic regions with speed and precision.
Related article: What is Genome Sequencing?- 3 Best Genome Sequencing Methods.
When resequencing is required?
Now, let’s see some of the applications of resequencing.
- It is used to report novel gene variants associated with a disease.
- It is used to fill gaps during whole-genome sequencing.
- It is used to understand or get insight into complex and repetitive genomic regions.
- It is used for the validation of pathogenic variants associated with a disease.
- It is used to study the coding regions of a gene.
- It is used for the validation of heterozygous variants.
- It is used to perform metagenomic analysis and to study unknown microbial populations.
- It is used to study the variants linked with mitochondrial DNA-associated diseases.
- It is used to resolve WGS complexity for a particular locus, region or gene.
- It is used to validate variants associated with rare genetic disorders.
Sequencing vs resequencing:
| Sequencing | Resequencing |
| Reading the order of nucleotides. | Reading the order of nucleotides that have been already sequenced. |
| Used to determine nucleotide sequence. | Used to validate variants found during targeted or WG sequencing. |
| Used to sequence a target region, gene or entire genome. | Used to study a specific sequence or region containing any important variants. |
| Used for research and diagnosis. | Particularly useful for diagnosis to report disease-associated variants. |
De novo sequencing vs resequencing:
| De novo sequencing | Resequencing |
| Refers to sequencing completely unknown regions. | Refers to sequencing the region that is already sequenced. |
| Doesn’t have a reference sequence to compare. | Do have a reference sequence or genome to compare. |
| Used to identify novel sequences, genes or genomic regions. | Used to validate disease-associated or pathogenic variants. |
| A DNA fragment is sequenced and assembled using a computational algorithm. | A DNA fragment is sequenced, compared with the reference sequence or genome and then resequenced to validate variants. |
| Extensively used in research. | Extensively used in clinical diagnosis. |
Related article: 47 Types of Sequencing Techniques You Should Know About.
Wrapping up:
In conclusion, resequencing is the most effective and necessary method for identifying sequence variations in clinical diagnosis. It provides scientists with clear and precise information about variants they have identified.
Notedly, state-of-the-art NGS platforms, available in recent times, don’t need resequencing in most of the cases. The readily available gene panels and disease assays are 99% accurate.
Still, scientists used to perform resequencing when they were not confident about the variant(s) they had identified.
Sources:
Resequencing Applications by ThermoFisher.
Targeted Resequencing by Illumina.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.