“Plant DNA extraction needs a CTAB DNA extraction buffer that contains ingredients that can effectively remove polysaccharides and polyphenols. Let’s see how it works and how to optimize the CTAB buffer for plant DNA extraction.”
DNA extraction is a process of isolating DNA from any biological cell to use it for analysis like PCR, sequencing or restriction digestion. Various chemical constituents of DNA extraction buffer allow effective isolation. But differences in the cell structure and chemical composition of a cell require a different DNA extraction setup.
For instance, the DNA extraction setup and buffer which works for animal tissue, can’t work for plants. In addition, plant DNA extraction is even a difficult task to execute because of their specialized cell structure and composition.
However, the CTAB-like buffer system can do better with such hard tissues. A micro-level optimization is required to use it for our use. Most of the time students fail to extract good DNA. There are two reasons for that.
First, they don’t know which buffer to use and second, they even don’t know how to use it. In this article, we will discuss plant DNA extraction and CTAB buffer. I will also explain variations and concentrations of different CTAB buffers that you can try and use.
Key Topics:
Plant DNA extraction:
Plants bear secondary metabolites and a diversely rigid cell wall made up of polysaccharides like pectin, cellulose, chitin, etc. In addition, the presence of turgor pressure, all these things creates problems in plant DNA extraction.
So the first thing one should have to keep in mind is that depending upon the constituent of a plant cell wall, and the type of polyphenols they contain, prepare a buffer system. I already have explained the different types of polysaccharides and polyphenols present in plants and required chemical modifications accordingly.
Go through this article: Why is DNA extraction from plants difficult?
Besides, the turgor pressure is another factor that we have to look after. Let me explain to you in brief, turgor pressure is water pressure that occurs from inside the cell to the cell wall. And eventually helps plants to stand straight.
Contrary, animal cells don’t have turgor pressure, as it is not actually required. And the soft membrane of the animal cells can’t bear it. So these three things (cell wall composition, secondary metabolites and turgor pressure) need advanced treatments.
A General DNA extraction scheme includes cell lysis, removal of contaminant and DNA stabilizing, precipitation and elution steps. In 1980, Murray and Thompson proposed an inexpensive and simple protocol for plant DNA extraction by CTAB. Dellaporta (1983) and Maroof et al., (1984) have optimized the existing protocols and developed excellent methods for plant DNA extraction.
Along with CTAB, chemicals like SDS and PVP gave an excellent yield for Corn and Soybeans. Such studies stamp CTAB as a major constituent of plant DNA extraction. Common steps in plant DNA extraction are,
Cell wall and nuclear membrane lysis- by chemical treatment, tissue homogenization using liquid nitrogen and physical grinding.
Removal of contaminants and other cell debris- by chemical treatment using SDS, CTAB, PVP, beta-mercaptoethanol and Triton X 100, etc.
Stabilizing and precipitating DNA- by chemical treatment using NaCl and alcohol.
Washing and elution- By alcohol and TE (Tris-EDTA) buffer.
How to extract DNA from plants?
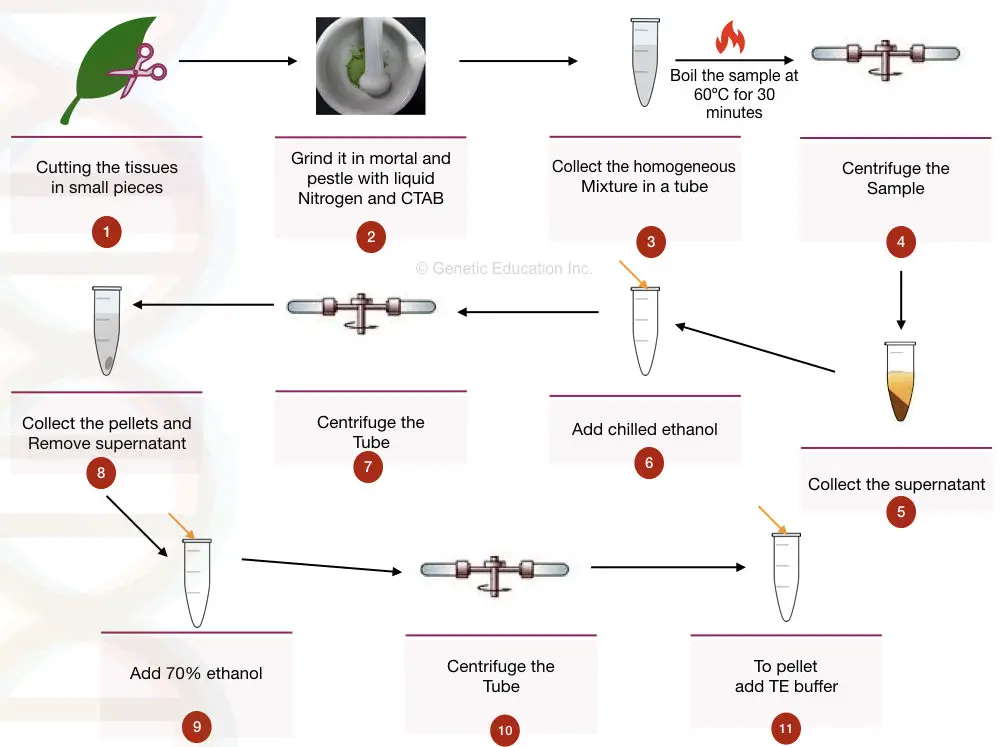
Select a plant species and prepare the protocol and chemicals accordingly. Start with sample preparation, clean the tissue and prepare it for homogenization.
Homogenization is a process to prepare a homogeneous mixture of plant tissue using tissue homogenization techniques like physical grinding using a Mortar and pestle.
Add liquid nitrogen and grind vigorously and add a lysis buffer, simultaneously. Liquid nitrogen prepares the fine power of tissue and also deactivates nucleases by providing an extra chill environment.
It instantly froze tissue below its freezing temperature and eventually ruptures the cell wall. However, care must be taken while handling liquid nitrogen. Take necessary precautions before.
Afterward, the homogenized sample would be incubated with lysis buffer and protease enzyme for effective lysis of the cell wall.
The nucleic acid is separated and precipitated using alcohol followed by a few rounds of washing with 70% ethanol.
The final precipitate would be eluted and dissolved in the TE buffer.
How to remove Tannin?
Tannin is actually the main culprit. It is a major constituent of polyphenolic compounds and is almost present in all plants. It is made up of a long phenolic ring and secreted during tissue damage, thus during the lysis process.
Previous studies demonstrated that Tannin is a proven PCR inhibitor that suppresses PCR amplification and remains bound with the DNA. Thus even if your DNA is good and your PCR fails repeatedly, you would have to worry about tannin.
PVP- PolyVinylPyrrolidone binds with the phenolic ring of tanning and prevents its interaction with DNA. In addition, the efficiency of PVP increases greatly by adding beta-mercaptoethanol.
The beta-mercaptoethanol remains in the supportive role, but effectively blocks the oxidation of tannin, thereby disallowing interaction with DNA. Conclusively, PVP and beta-mercaptoethanol must be there in the plant DNA extraction buffer with the CTAB.
Now let us come to our major player- CTAB.
CTAB in plant DNA extraction:
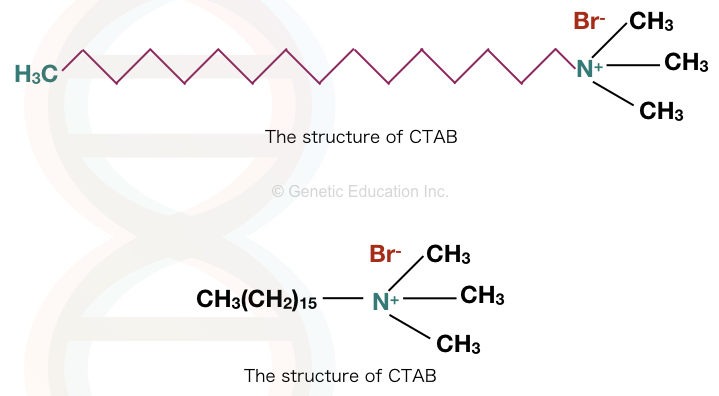
CTAB, cetyltrimethylammonium bromide, also called Cetrimonium bromide or hexadecyltrimethylammonium bromide. The molecular formula of CTAB is [(C16H33)N(CH3)3]Br.
It is a cationic detergent, and water and alcohol soluble. Care must be taken while handling CTAB as it has corrosive and toxic properties. The structure is explained in the figure below.
Besides its role in DNA extraction, CTAB is used as an antiseptic (against bacteria and fungus), detergent and ingredient of cosmetics.
In principle, in the extraction process, CTAB associates with the cell wall polysaccharides and proteins and co-precipitates it along with DNA. By centrifugation, such co-precipitates can be removed in consecutive steps.
“Do you know the CTAB can also be used in the SDS-PAGE?
The positively charged CTAB can readily bind with the negatively charged glycoproteins during protein electrophoresis. Instead of fuzzy bands, clear and sharp bands can be achieved by using a pinch of CTAB into the SDS PAGE. “
CTAB DNA extraction buffer:
Major ingredients of CTAB buffer are CTAB, Tris, EDTA, beta-mercaptoethanol, PVP and NaCl salt. Other chemicals required in the process are SDS, protease enzyme, Phenol, chloroform, isoamyl alcohol and ethanol.
Notedly, keep in mind that the composition of the buffer and concentration of chemicals would differ from plant species to species.
10% CTAB solution:
- CTAB- 100g/1000ml
- 0.7M NaCl- 41g/1000ml
- dH2O- up to 1000ml
2% CTAB solution:
- 2% CTAB- 20g/1000ml
- 0.7M NaCl- 41 g/1000ml
- dH2O- up to 1000ml.
Major ingredients of CTAB buffer:
| Chemical | Required concentration |
| CTAB | 2% (w/v) |
| Tris (pH 8.0) | 100mM |
| EDTA | 20mM |
| NaCl | 1.5M |
| PVP | 1 to 2% (w/v) |
| Beta-mercaptoethanol | 0.2% |
2X CTAB buffer preparation
- 2% CTAB solution- 20g
- 1.4M NaCl- 81.8g
- 100mM Tris (pH 8.0)- 100ml
- 20mM EDTA- 40ml
- 2% PVP- 5g
- ~0.4% beta-mercaptoethanol- 4ml
- Final volume- 1000ml with dH2O
| Components | 10X stock solution | 2X working solution | Volume for 1000mL |
| CTAB solution | 10% | 2% | 20ml |
| NaCl | 7M | 1.4M | 81.8gm |
| Tris | 500mM | 100mM | 100ml |
| EDTA | 100mM | 20mM | 40ml |
| PVP | 10% | 2% | 10gm |
| Mercaptoethanol | 2% | 0.4% (roughly) | Add just before use |
| D/W | – | Up to 1000ml final volume |
- Pre-prepare a CTAB solution (10%) by heating and store it at 37°C temperature. Prepare a working solution from stock. Avoid using 2 or more days-old CTAB solutions.
- Add the beta-mercaptoethanol just before the use.
- Prepare a 10X stock for CTAB solution to avoid repeated chemical preparation.
- Also, prepare a 10% stock solution of PVP.
- Store beta-mercaptoethanol at 4°C
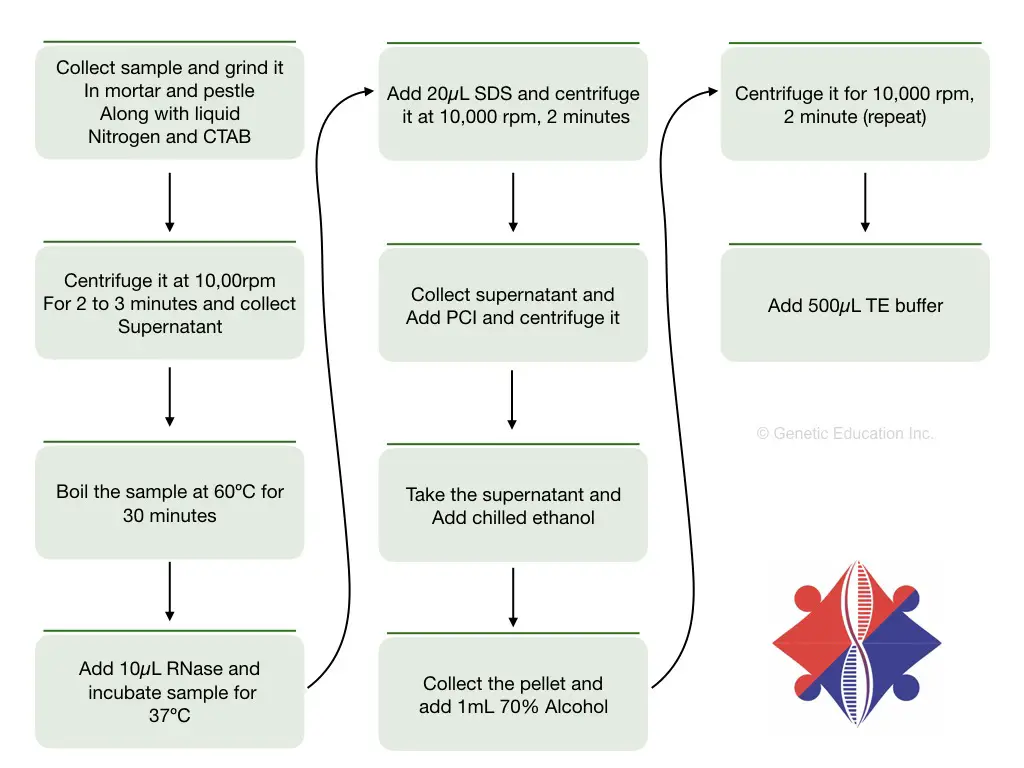
CTAB plant DNA extraction protocol:
Plant DNA extraction using CTAB is simple and effective. However, other chemicals like RNase, proteinase K, SDS and PCI may be used to improve the quality and yield of DNA.
This protocol is my own, we used it in our labs during our masters. Let us see the protocol.
- Take 5gms of fresh plant tissue and cut it into small pieces.
- Add liquid nitrogen to the tissue and roughly grind the sample into the mortar and pestle.
- After the tissue becomes a powder, add 500 μL of CTAB extraction buffer and beta-mercaptoethanol, grind it again followed by vortexing for 5 minutes.
- After it becomes homogeneous, take the mixture into the 2ml Eppendorf tube.
- Boil the sample for 30 minutes at 60°C to 65°C.
- Centrifuge the sample at 25,000rpm for 5 to 8 minutes and transfer the supernatant into another tube.
- Be careful, take the only supernatant, and don’t mix the supernatant with the debris.
- Now add 5 μL to 10 μL of RNase solution to the supernatant and incubate at 37°C for 25 minutes.
- The proteinase K step is additional (you can use it if necessary).
- Centrifuge the sample at 10,000rpm for 2 minutes and take supernatant to another tube.
- Now add chilled isopropanol (70%), and a pinch of NaCl to the supernatant and precipitate the DNA by inverting the tube several times.
- Centrifuge the precipitate for 10,000rpm for 2 minutes and collect the pellet (remove the supernatant).
- Now wash the DNA pellet with alcohol two times at 9000 rpm for 2 minutes.
- After the clear pellet appears, add 500μL of TE buffer or elution buffer to the pellet and dissolve the DNA in it.
Bonus tips
If DNA precipitates are not observed after the addition of the chilled alcohol, then place the tube at -20°C for 15 to 20 minutes and re-precipitate it.
Read more:
- 10 Secret Tips for DNA Extraction to Get Good Results
- How to Prepare Lysis Buffer for Different Types of DNA Extraction Methods?
Optimizations and modification:
No universal CTAB DNA extraction protocol exists, keep in mind this thing. If someone says it, it’s wrong. You have to modify the protocol and preparation as per your need.
For example, some plants need only simple grinding in mortar and pestle, while some need liquid nitrogen. Plenty of proven DNA extraction chemicals are available, you can choose according to your need.
SDS (sodium dodecyl sulfate) is an excellent anionic detergent that can lyse proteins. Edwards et al. (1991) used 0.5% SDS with the CTAB buffer and developed a simple plant DNA extraction protocol.
Proteinase K is yet another excellent ingredient that also can digest proteins- DNase and RNase present in the cell and protects the nucleic acid from degradation. Use proteinase K overnight at room temperature or for 2 hours at 60°C.
To make things even more effective use RNase as well in combination with proteinase K. Proteinase K and RNase are commercially available. Use the manufacturer’s guide for use.
Use PCI (phenol: chloroform: isoamyl alcohol) in combination with the CTAB buffer. Treat the sample with PCI, centrifuge it, and collect the supernatant part.
Follow this protocol on how to use PCI: Phenol, chloroform and isoamyl alcohol DNA extraction.
Autoclave all the utilities prior to use including solutions. Do not autoclave alcohol and phenol.
Urea is yet another excellent lysis agent that can be used along and alone in DNA extraction.
Use of Urea in plant DNA extraction:
- Grind the tissue in a mortar and pestle and add Urea buffer (which contains Urea, Tris, EDTA and NaCl).
- Add liquid nitrogen and homogenize it.
- Take a sample in the Eppendorf tube and centrifuge it at 12,000rpm for 2 minutes.
- Discard pellet and take supernatant in another tube
- Add chilled ethanol or ammonium acetate
- Wash the sample with the alcohol (minimum 2 wash)
- Dissolve the DNA in 500μL of TE buffer.
The use of Urea is the easy and safest way to extract DNA from plants and sufficient enough for routine PCR and restriction digestion but not enough for sequencing or microarray analysis.
Bonus tips
The ‘mantra’ to success in plant DNA extraction is to grind tissue well, hard, and rough until fine homogenization. This will increase the yield.
Advantages:
- CTAB DNA extraction is a simple, effective, cheap and reliable method to extract DNA from plants, especially for RAPD analysis and barcoding.
Disadvantages:
- CTAB method is an organic solvent-based extraction method that has health hazards and harmful properties.
- CTAB is unsafe and hazardous to use.
- Liquid nitrogen can burn the skill and even do permanent damage.
- Phenol is volatile and can burn the skin.
- Beta-mercaptoethanol can irritate upon inhalation.
Wrapping up:
Plant DNA extraction is a hard, tedious and time-consuming process. It actually challenges our knowledge to optimize things in our lab. However, the use of a CTAB buffer will surely help in the majority of cases.
Nonetheless, to get success every time in plant DNA extraction, the first thing is to understand the structure and constituent of the plant cell of the species you are using. It will surely make things easy.
Importantly, follow a common lab safely and perform the experiments using liquid nitrogen under the supervision of experts.