“A method used to detect a single base change or SNP using sequence-specific primers is referred to as ARMS or Allele-specific PCR.”
The PCR technique can detect mutations like deletion, duplications, insertion or single base change- SNP. It’s a temperature-dependent amplification technique that relies on Taq DNA polymerase.
Many variations of PCR exist depending upon the assay requirement, ARMS-PCR is one among them. It has other names like allele-specific PCR, PASA or AS- PCR, all have similar applications.
Let us know the full names, ARMS acronym as Amplification Refractory Mutation System. PASA acronyms as PCR Amplification of Specific Allele and AS- PCR stand for Allele-Specific PCR. It uses the word “Allele” many times, the reason is its specific amplification capacity for a specific allele or DNA sequence.
Kary Mullis described the technique of in vitro amplification in the year 1983. After a few years of the discovery of the actual PCR technique, C. R. Newton and coworkers discovered the ARMS-PCR or allele-specific PCR technique.
The present technique has applications in detecting SNPs (Single nucleotide polymorphism) and genotyping. That’s all we are going to discuss in the present article.
The present post comprises information on ARMS- PCR, its principle, process, protocol and steps. I have also explained the advantages, disadvantages and applications. This article will boost your PCR knowledge and will surely help you in your study.
Stay tuned.
Key Topics:
What is ARMS or Allele-Specific PCR?
Alleles are alternative gene forms. When a mutation occurs, it produces two different alleles for a gene; one mutant one and another normal one. The Allele-specific PCR has the power to detect a single specific allele.
Meaning, If you wish to amplify only a mutant allele, design a primer set accordingly and amplify it using this technique. Each set of specific primers is designed for each specific allele.
However, we need to do a minor change in primers to amplify each allele. PCR amplifies each allele simultaneously, afterward. While gel electrophoresis prepares results to evaluate.
Two different primer sets are prepared for separate alleles. The mutant allele-specific primers are refractory (resistant) to normal allele and vice versa. Henceforth, known as Amplification Refractory Mutation System. R. Newton had given the name ARMS- PCR.
Principle of ARMS- PCR:
The principle of the present technique relies on the modification of primers to amplify a specific allele.
The 3’ end of the primer is modified in such a way that one primer can amplify a mutant allele while the other can amplify the normal allele. To do this, researchers modify a few bases from the primer’s 3’ OH end.
Only a single set of primers can only amplify a specific allele present in a sample, during the reaction. Gel electrophoresis helps to examine results.
The mismatch concept:
By introducing a mismatch in a primer makes refractory amplification possible. Here the mismatch between the primer and the template DNA plays a crucial role in achieving the amplification.
Mismatch indeed alters the annealing temperature for different sets of primers. Here are examples of several mismatches.
Strong mismatch: G/A, C/T, T/T
Medium mismatch: A/A, G/G, C/C,
Weak mismatch: C/A, G/T
Think about this question, I will answer it later. Why aren’t we using high-fidelity Taq DNA polymerase in ARMS PCR?
See the image below so you can understand the concept easily.
Procedure and steps of ARMS-PCR:
The process is simple and effective. Hazardous radiolabeled probes aren’t involved here. We can divide the whole process into 4 separate steps:
- Primer designing
- Amplification
- Agarose gel electrophoresis
- Results interpretations
Primer designing:
Primer preparation and designing play a crucial role here. Remember the primer must be allele-specific. A few points to keep in mind while preparing are,
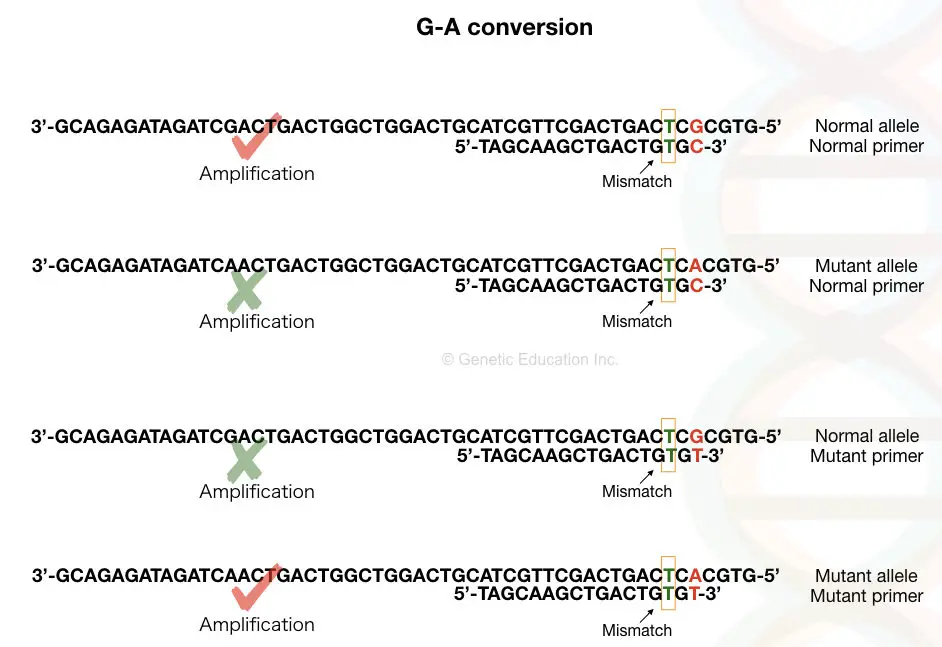
See the image above first, Suppose our DNA sequence has G-A point mutation viz, G in normal allele and A in place of G in the mutant allele.
We have to design a forward primer in such a manner that for the normal allele the primer contains C (complementary to G) at 3’ end and the mutant primer contains T in place of C.
Although it works magically when we add a mismatched base near our SNP at the 3’ end. You may wonder why we need to add a mismatch? The mismatch is the key factor in achieving the amplification.
Weak mismatch elevates chances of amplification and so add strong mismatch to avoid false-amplification near the 3’-OH end of the primer (at -2 position ideally). Mismatch prevents binding to the non-complementary sequence and terminates amplification.
C:T, G:A and A:G is the strong mismatch base pairs that reduce the amplification process up to 100 fold. We have to modify only a single primer, either forward or reverse, the other primer usually remains unchanged and can work for either allele.
Besides, the primer should follow all the standard criteria for primer designing. It must have less GC content, less hairpin structure and a length between 20 to 30 nucleotides.
Amplification:
Usually, we do not discuss the amplification process separately. But in order to get ‘correct’ amplification, we should have to maintain healthy amplification conditions. Here are a few,
Set higher annealing temperature, depending upon the primer’s highest annealing temperature. If set lower, it increases the chances of non-specific amplification, remember, we are playing with only a single base change primer.
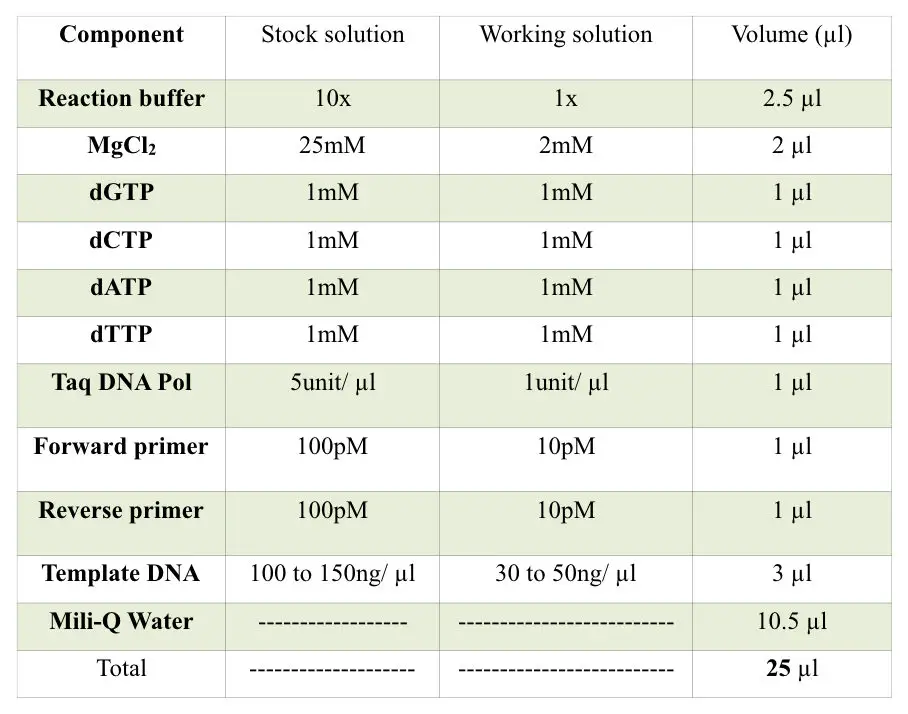
Prepare reaction following the table below.
Set a lower PCR cycle to maintain the specificity of the reaction. Ideally set 22 to 25 cycles only. More PCR cycles increase the chances of non-specific bindings, primer-dimer and redundant results. It gives false-positive results.
“An additional mismatch to the 3’ end after the first mismatch increases the specificity of the ARMS-PCR reaction.”
Use a few internal controls to monitor results. Include negative, positive and internal control tubes in reaction.
Agarose gel electrophoresis:
The ARMS PCR has direct diagnostic use, the reason is that it doesn’t require hybridization steps. The results are run on 2% agarose gel, ideally under proper agarose gel running conditions. Along with, add a 1000 bp molecular marker.
The gel electrophoresis technique is sufficient to separate different alleles and evaluate results. ARMS- PCR results are interesting, go to the next section.
Results and interpretation:
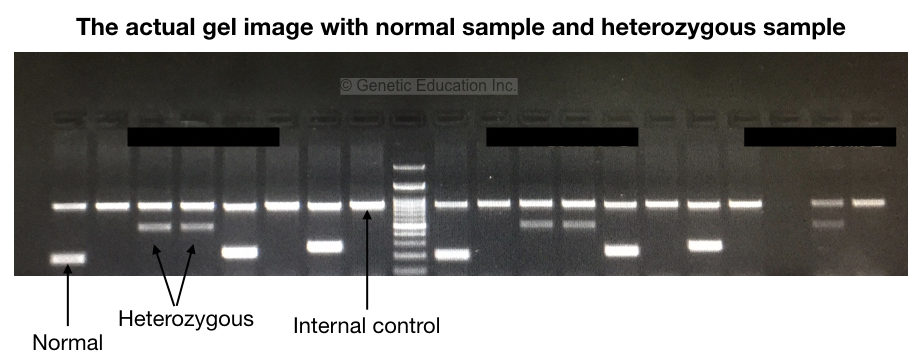
To examine results, the very first is to observe the internal control. The internal control band must be present in all reactions. It shows that our reaction preparation, cycling conditions and other practices are fine.
We have shown results in the gel image given above. Please first refer to it. Now analyze each band for the normal and mutant alleles. To understand the results let’s prepare a hypothetical situation.
Suppose we have three samples; one normal, one heterozygous carrier and one homozygous dominant. We have prepared 2 tubes for each sample, a total of 6 tubes for samples with one positive control and two negative controls.
See the figure,
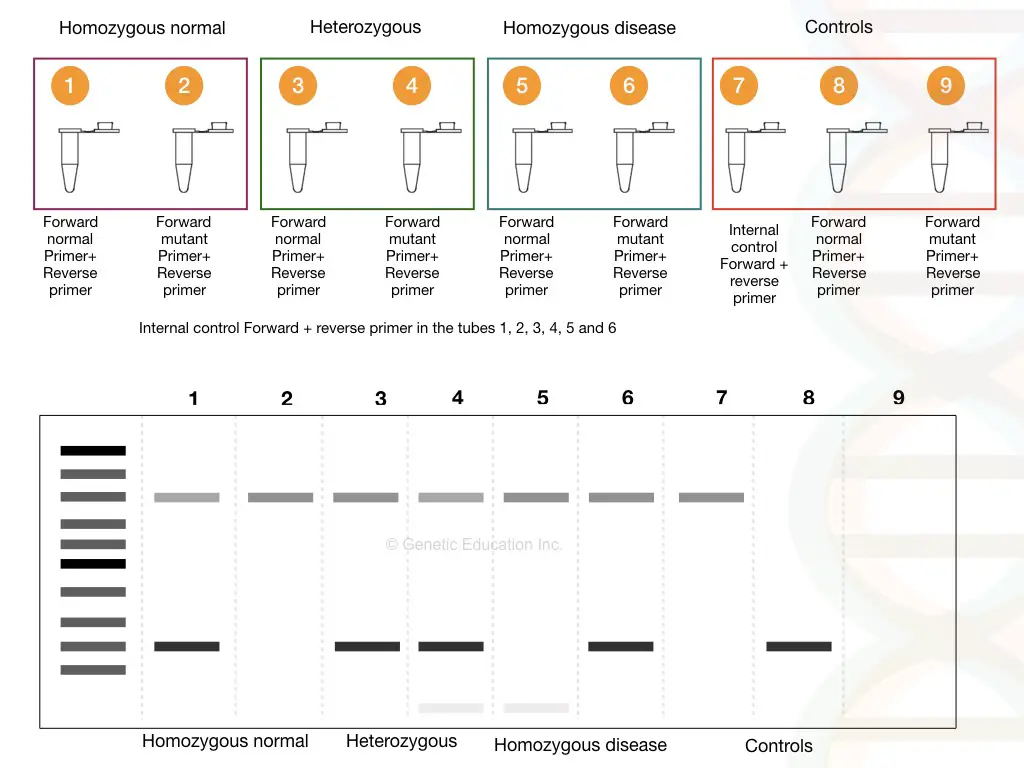
As shown in the figure above, homozygous normal shows a single DNA band, heterozygous carrier shows two bands (one for normal and one for mutant allele) and homozygous disease conditions show a single mutant band.
It can also be stated like this, the mutant primer can’t amplify the normal homozygous allele, both primers amplify both alleles in heterozygous and only a single mutant primer can amplify the mutant allele.
Advantages of ARMS-PCR:
Every PCR variant has its own importance and so does the allele-specific PCR too. Here I have listed some advantages of it.
- The technique can distinguish two different alleles precisely.
- It can detect a single base variation- SNP.
- It’s an accurate, fast and reliable PCR technique.
Limitations of ARMS-PCR:
- It includes complex techniques of primer designing and incorporating mismatches.
- It can detect only a few SNPs at a time.
- It can precisely distinguish homozygous and heterozygous alleles.
- It is one of the most important techniques for genotyping and allelic variation studies.
- It includes more primer sets hence is a costly process.
- It can detect only known SNP only, new variation or mutation can’t be detected.
- It has higher chances of false-positive results and that’s why every time it needs internal, negative and positive controls.
- It’s temperature-sensitive, meaning, a minute alteration in annealing temperature causes a serious problem in the reaction.
- Lastly, much like other PCR variants, it can’t examine chromosomal alterations, larger mutations like deletions and duplications.
Applications of ARMS-PCR:
- The ARMS-PCR technique has exclusive application in SNP detection, genotyping and finding allelic variation.
- It often applies in the detection of various mutations.
- It has applications in diagnostic too. For example, it detects different alleles of Beta-thalassemia or mutations using sequence-specific primer sets. It’s a gold standard method in detecting mutations associated with Beta-thalassemia and sickle cell anemia.
- Researchers also use allele-specific PCR in detecting JAK2 mutations and HIV variants. Commercial kits are now available for various assays. Knok and et al have used this technique in HIV- 1 genotyping for the ATT gene deficiency. Read their research here, Click here.
Optimization for ARMS-PCR:
Our blog is not just about gathering and providing information present on the internet. We also share our own experiences too. I have a huge experience in PCR technology and that’s why we add more value to the topic.
In this section, I will give you some optimization tips that help not only students but also newbie researchers in their experiments. Here are my suggestions,
- Avoid longer primers for ARMS-PCR, use a primer length between 25 to 28 nucleotides.
- Incorporate a single mismatch at the 2nd position at the 3-OH end.
- Add a second/ weak mismatch at the 3rd position.
- Use 10pM each ARMS primer, use 1 to 3pM internal primer.
- Use shorter PCR programs usually, between 25 to 28.
- Use MgCl2 to the reaction to increase the specificity of amplification. Ideally use > 1.5mM.
Remember the question I had asked at starting? Why aren’t we using high-fidelity DNA polymerase in ARMS-PCR? Here is the answer,
In the conventional ARMS-PCR, we use only a simple Taq DNA polymerase. The high-fidelity DNA polymerase has polymerase as well as exonuclease activity and hence will remove the mismatches.
Interesting information: “Guanine” to other nucleotide conversion is more prevalent than rest.
Variants of ARMS-PCR:
Scientists do modify the native ARMS-PCR technique to perform various functions. Here are some of the modifications.
Concept of T-ARMS-PCR:
The T-ARMS-PCR termed as tetra-primer PCR or tetra-ARMS-PCR is used in detecting various mutations and hence has great diagnostic values. It uses 4 different primers for two separate alleles of a gene.
Meaning a set of forward and reverse primer for a normal allele and another set of forward and reverse primer for a mutant allele.
Multiplex- ARMS PCR:
Multiplex-ARMS PCR is used for a similar purpose, however, it can save time and cost of the experiment. Here, two separate multiplex reactions have been prepared for more than two SNP detection.
It’s more complex and elevates chances of false results. One needs substantial experience in the relevant field to design the assay and to execute results.
qARMS-PCR:
It is a technique that not only amplifies alleles but also quantifies the specific allele. Henceforth, it can tell us the amount of mutant or normal alleles present in the sample.
Wrapping up:
To understand the concept of Allele-specific PCR or ARMS-PCR, one should have a strong experimental background. At least, they know various types. It is very difficult for a newbie or a student to understand the concept.
Because the concept is a bit complicated. We are doing serious changes to primers, we usually aren’t recommended.
The technique has a great utilization in detecting SNP, genotyping and mutation and I personally have designed it and used it during my research period.
Trust me it was fun to do trial and error. I hope you may like this article. If yes, please Bookmark it on your browser.
Sources:
Darawi, M.N., Ai-Vyrn, C., Ramasamy, K. et al. Allele-specific polymerase chain reaction for the detection of Alzheimer’s disease-related single nucleotide polymorphisms. BMC Med Genet 14, 27 (2013). https://doi.org/10.1186/1471-2350-14-27.
Gaudet M, Fara AG, Beritognolo I, Sabatti M. Allele-specific PCR in SNP genotyping. Methods Mol Biol. 2009;578:415-24. doi: 10.1007/978-1-60327-411-1_26. PMID: 19768609.