“Here is the list and explanation of 10 crucial information revealed by ancient DNA with examples.”
Ancient DNA is the DNA obtained from ancient remains or fossils. However, it can not be obtained as a whole! But the fragments that we get from such ‘remains’ reveal crucial information regarding life on Earth.
We get lucrative knowledge regarding the life of that time, their population, genetic diversity, special traits, limitations, their interaction with the ecosystem and how they extinct.
Just a tiny string of DNA gives us such information and allows us to understand the life and biodiversity of that particular time. It’s like a tiny time capsule, using which, we can explore that particular time, where the ancient DNA belongs!
Sounds amazing!
Let’s learn what crucial information ancient DNA reveals in this article. I will explain every point with an example and will make it more understandable.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Related article: Gene Flow vs. Genetic Drift- The Battle Of Genetic Forces.
Key Topics:
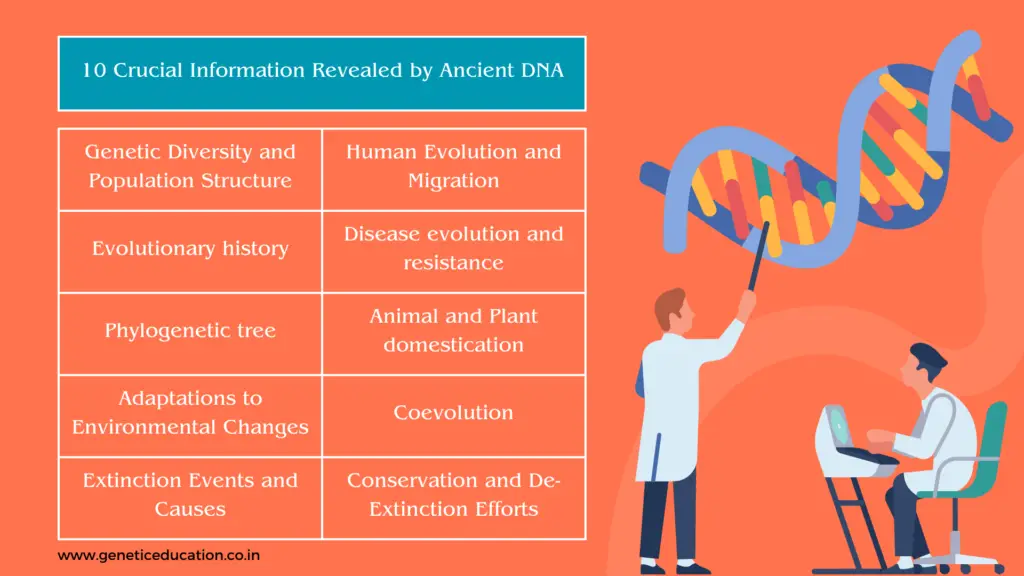
10 Crucial information revealed by ancient DNA
Ancient DNA holds information on genetic diversity and population, Evolutionary history, phylogenetic structure, adaptation to environmental changes, Extinction events and causes, Human evolution and migration, disease evolution and resistance, animal and plant domestication, coevolution, species distribution, conservation and de-extinction events.
We will discuss every point elaboratively here.
Genetic Diversity and Population Structure:
Ancient DNA study allows scientists to understand the genetic diversity and population structure of the organism to which the DNA belongs, which once roamed on Earth. Genetic diversity simply means the diversity in the gene pool.
The more diverse the genes, the better a population can adapt to changing environments, diseases, and other challenges. By looking into how diverse and adaptive genes they have, scientists can not only understand their genetic richness but also their population structure.
Population structure means how different species of a particular population to which the DNA belongs are connected. Migration patterns and evolution can be tracked through it.
A study published in the Journal of Current Biology by Skoglund et al. Examined the ancient DNA of North American gray Wolf. The ancient DNA sample of a gray Wolf that scientists have is ~50,000 years old.
By looking at their DNA scientists can now understand how they could adapt to specific environments and what genetic factors gave adaptability to some wolves hunting in forests while others in open landscapes.
In addition to this, population structural analysis reveals how these wolf populations were distributed across North America.
Interestingly, by analyzing the ancient DNA obtained from various wolf species across North America– Rocky Mountains, the Arctic tundra and the woodlands of the East, scientists can establish the common and distinct link between these populations.
Thereby they reconstructed the historical dynamics of wolf populations and what forces forced them to migrate to different regions.
Evolutionary history:
Consider our evolution as a giant storybook. The whole storybook is interconnected with chapters, without which it’s incomplete. Unfortunately, we lack many chapters and links to our evolutionary history.
Many theories and hypotheses we have on our evolution. However, every theory has loopholes and lacks a concrete foundation. By looking into the ancient remains, either of bacteria or any other eukaryote, scientists can understand the evolutionary dynamics.
A study published in the journal PNAS by Librado et al. (2015) explained the genetic basis of how the Yakutian horses adapted to the subarctic environment. The evolution of extremely hairy coats and climate-specific metabolic activities helped them survive in extremely cold conditions.
Another study published in Oxford Academics by McHorse B et al. (2019) showed that the sleek and single-toed horses that we know today evolved from multi-toed horses.
Phylogenetic tree:
Here comes the next point, what next after knowing evolution? To know how each species and organism are interlinked. Phylogenetics is the study to understand how different species or organisms are connected.
Let’s understand it through the example of a great Dinosaur Family. Scientists now know that different dinosaur species lived in different parts of the world.
Analysis of various ancient dinosaur DNA samples helps prepare relationships between species and samples obtained from different regions of the world. Using this information scientists prepared a relationship tree of closely and distinctly related species.
A study published in the Journals of Systematic Paleontology by Langer & Banton (2010) explained the phylogenetic tree structure of dinosaurs and showed closely and distinctly related species.
This helps understand the reason for diversity, migration, competition with other species, and extinction of dinosaurs.
Related article: What is a Phylogenetic Tree and How to Construct it?
Adaptations to Environmental Changes:
Is it possible to forecast the environment or climate thousands to millions of years ago by just looking into the ancient string of DNA? First, take a look at the example of Arctic Foxes.
By studying ancient DNA from Arctic foxes, scientists can understand how these furry creatures adapted to the “freezing” cold over time. Maybe they developed thicker fur or changed their hunting habits.
By looking into an organism’s genes, scientists can understand why those genes evolved and for which function. Using that information, scientists can forecast the climate conditions or environmental factors that exist.
Extinction Events and Causes:
Ancient DNA studies also reveal the fact that different species disappeared from Earth due to environmental pressure and human interference. Let’s take the example of giant, wooly and colossal mammoths.
A study published in the Journal of Current Biology by Palkopoulou et al. (2015) explained the genetic decline in context to the extinction of mammoths. Their findings showed that loss of genetic diversity, inbreeding, climate change and loss of suitable habitats are the major causes of their extinction.
Human Evolution and Migration
Ancient DNA is a great tool to understand human evolution and migration. The movement of early human populations, interactions with other hominid and hominin species, and the development of different human lineages can be tracked accurately.
A study published in Nature by Skoglund et al. (2016) revealed the migration pattern of modern humans and their migration from South Africa to the South West Pacific Region.
By studying genetic markers they tracked the migration root, enabled reconstructing it, and understood the population diversification, migration needs and adaptation dynamics in new habitats.
In addition, several mitochondrial DNA studies helped to piece together the story of human evolution and migration and understand how our ancestry dispersed across continents.
Disease evolution and resistance:
The Human race faced many lethal infectious outbreaks in the past. Although resistance was developed and spread over time, millions of people died by various infections (which are irrelevant in the present time).
Analysis of ancient DNA obtained from the remains of the infected patients manifests crucial information on disease or infection evolution, and resistance development. Consider the infamous black death by Plague.
A study published in PLOS ONE by Bos et al (2010) unfolds the mystery of the black death during the Middle Ages in Europe. They identified the responsible strain Yersinia pestis and understood the evolution and virulence factors that contributed to the infection.
Further to this, they also showed how the human population had developed a resistance mechanism overtime against the pathogen. This shows how crucial the ancient DNA is as evidence for infection and resistance studies.
Animal and Plant domestication:
Ancient DNA provides a sheer genetic proof of animal and plant domestication. It explains what genetic changes in their genome helped in their domestication.
A study published in the Cell Reports by Parker et al. (2017) studied how wild wolves converted into domestic animals and what genetic, environmental and geographical changes drove the conversion.
Their comprehensive analysis revealed that variations in genes associated with behavior, appearance, and digestion are the key genetic factors in this process. Many other studies on other domestic animals and plants also support the present findings.
Coevolution:
Coevolution is a process in which two separate organisms affect each other’s evolution process through natural selection. The genetic interactions between hosts and pathogens have shaped immune responses and the resistance development as well as the synthesis of a new set of genes in the host and pathogen, respectively.
A study published in MDPI by Pimenoff et al. (2018) showed the coevolution of HPV. It explains the strategies and genetic changes developed by both humans and pathogens for coevolution.
Conservation and De-Extinction Efforts:
Data and research provided by the ancient DNA studies are pivotal for our de-extinction efforts and bio-conservation. By studying the past— the genetic composition and environmental factors, we can protect the present.
For example, by analyzing the historical genetic information for one particular animal (which is on the verge of extinction), we can prepare strategies to conserve them. These data provide insight into what adverse factors lead to the reduction in their population size and how we can manage them.
Research published in the WILEY online library by Leonard (2008) showed how ancient DNA helped in the conservation of various species with examples. You can read this paper to learn more about this sub-topic.
Read more: Importance of Molecular Phylogeny.
Wrapping up:
Ancient DNA is a valuable asset we obtained from biological remains. It’s like a time machine for us and helps us revisit the time that it belongs. However, the major problem is the yield of DNA.
The DNA obtained from ancient remains isn’t intact and sufficient. Thus, we can’t unfold the complete story. However, still, those strings left and survived for millions of years, and adverse climates are valuable assets for the scientific community.
I hope this article helped you and got you interested in ancient DNA studies. If so, do share this blog post with your friends and on your social media.
Sources:
- Skoglund P, Ersmark E, Palkopoulou E, Dalén L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr Biol. 2015 Jun 1;25(11):1515-9. doi: 10.1016/j.cub.2015.04.019. Epub 2015 May 21. PMID: 26004765.
- Librado P, Der Sarkissian C, Ermini L, Schubert M, Jónsson H, Albrechtsen A, Fumagalli M, Yang MA, Gamba C, Seguin-Orlando A, Mortensen CD, Petersen B, Hoover CA, Lorente-Galdos B, Nedoluzhko A, Boulygina E, Tsygankova S, Neuditschko M, Jagannathan V, Thèves C, Alfarhan AH, Alquraishi SA, Al-Rasheid KA, Sicheritz-Ponten T, Popov R, Grigoriev S, Alekseev AN, Rubin EM, McCue M, Rieder S, Leeb T, Tikhonov A, Crubézy E, Slatkin M, Marques-Bonet T, Nielsen R, Willerslev E, Kantanen J, Prokhortchouk E, Orlando L. Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc Natl Acad Sci U S A. 2015 Dec 15;112(50):E6889-97. doi: 10.1073/pnas.1513696112. Epub 2015 Nov 23. PMID: 26598656; PMCID: PMC4687531.
- McHorse, Brianna K., Andrew A. Biewener, and Stephanie E. Pierce. “The Evolution of a Single Toe in Horses: Causes, Consequences, and the Way Forward.” Integrative and Comparative Biology 59, no. 3 (2019): 638-655. Accessed December 17, 2023. https://doi.org/10.1093/icb/icz050.
- Max C. Langer & Michael J. Benton (2006) Early dinosaurs: A phylogenetic study, Journal of Systematic Paleontology, 4:4, 309-358, DOI: 10.1017/S1477201906001970.
- Palkopoulou E, Mallick S, Skoglund P, Enk J, Rohland N, Li H, Omrak A, Vartanyan S, Poinar H, Götherström A, Reich D, Dalén L. Complete genomes reveal signatures of demographic and genetic declines in the wooly mammoth. Curr Biol. 2015 May 18;25(10):1395-400. doi: 10.1016/j.cub.2015.04.007. Epub 2015 Apr 23. PMID: 25913407; PMCID: PMC4439331.
- Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, Fu Q, Harney E, Lipson M, Mallick S, Novak M, Rohland N, Stewardson K, Abdullah S, Cox MP, Friedlaender FR, Friedlaender JS, Kivisild T, Koki G, Kusuma P, Merriwether DA, Ricaut FX, Wee JT, Patterson N, Krause J, Pinhasi R, Reich D. Genomic insights into the peopling of the Southwest Pacific. Nature. 2016 Oct 27;538(7626):510-513. doi: 10.1038/nature19844. Epub 2016 Oct 3. PMID: 27698418; PMCID: PMC5515717.
- Bos, Kirsten I., Philip Stevens, Kay Nieselt, Hendrik N. Poinar, Sharon N. DeWitte, and Johannes Krause. “Yersinia Pestis: New Evidence for an Old Infection.” PLOS ONE 7, no. 11 (2012): e49803. Accessed December 17, 2023. https://doi.org/10.1371/journal.pone.0049803.
- Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, Ostrander EA. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development. Cell Rep. 2017 Apr 25;19(4):697-708. doi: 10.1016/j.celrep.2017.03.079. PMID: 28445722; PMCID: PMC5492993.
- LEONARD, JENNIFER A. “Ancient DNA Applications for Wildlife Conservation.” Molecular Ecology 17, no. 19 (2008): 4186-4196. Accessed December 17, 2023. https://doi.org/10.1111/j.1365-294X.2008.03891.x.
- Pimenoff VN, Houldcroft CJ, Rifkin RF, Underdown S. The Role of aDNA in Understanding the Coevolutionary Patterns of Human Sexually Transmitted Infections. Genes (Basel). 2018 June 25;9(7):317. doi: 10.3390/genes9070317. PMID: 29941858; PMCID: PMC6070984.

