“In this article, Learn about various SNP genotyping techniques using PCR, microarray and sequencing-based approaches.”
SNP (Single Nucleotide Polymorphism) is a single base alteration in a DNA, gene or sequence. An SNP may alter a genetic code and consequence in an altered phenotype if occurred in a coding sequence.
Furthermore, reports also suggest that several SNPs in non-coding and promoter regions showed similar actions. Thus, studying SNPs can help understand the pathogenicity of an individual SNP or group of SNPs on an individual’s or population’s health.
The SNP detection technique, popularly known as “SNP genotyping”, helps distinguish the homozygous and heterozygous conditions associated with the SNP.
In this article, I will explain to you several techniques that scientists use for SNP genotyping. The present article will help beginner students understand the basics of SNP genotyping while researchers choose the most suitable technique for their research.
Stay tuned.
Key Topics:
10 SNP Genotyping Techniques
For deeper understanding and learning feasibility, I have divided the present guide into three parts viz. PCR-based genotyping, array-based genotyping and sequencing-based genotyping.
- PCR based techniques
- ARMS PCR
- RFLP analysis
- TaqMan PCR analysis
- High-resolution melting curve analysis
- KASP
- Digital PCR
- Microarray-based techniques
- SNP microarray
- Sequencing-based genotyping
- Sanger or targeted sequencing
- NGS-base genotyping
PCR-based SNP genotyping
In the present method, PCR amplification is used along with subsidiary techniques like restriction digestion, agarose gel electrophoresis and probe-hybridization. It’s a low Throughput approach in which we can only determine a few SNPs.
ARMS-PCR:
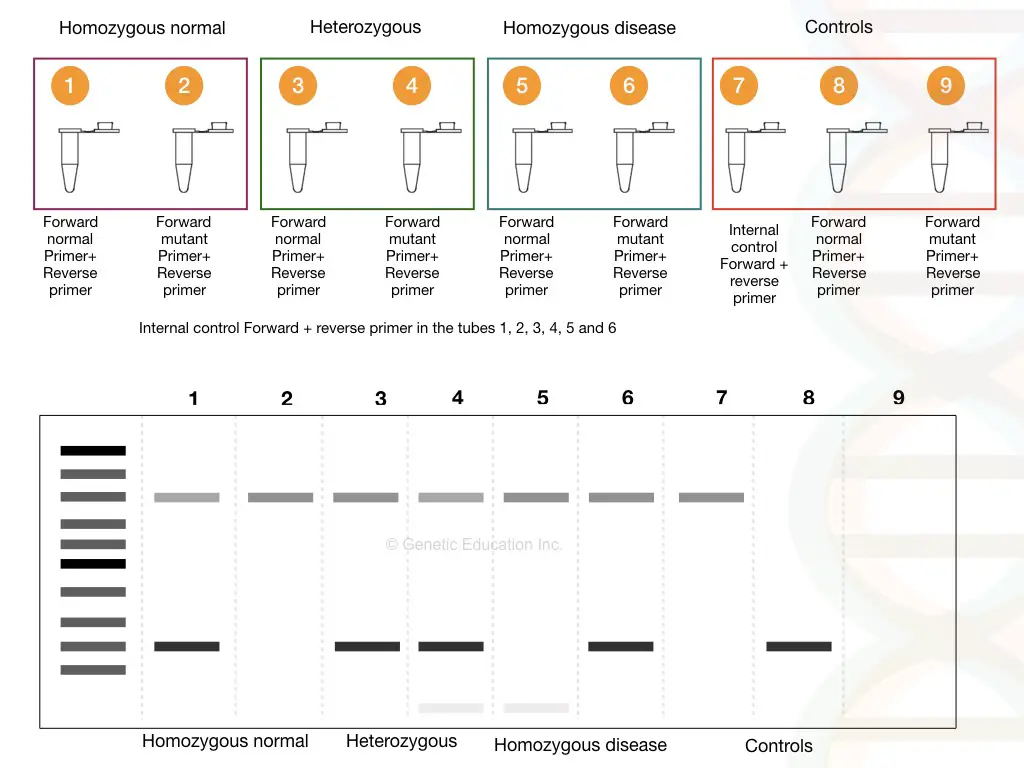
ARMS (Amplification Refractory Mutations System) or Allele-Specific PCR is an agarose gel electrophoresis-based technique used to selectively amplify the SNP-containing allele.
Here the SNP-containing allele is selectively amplified by designing ARMS-Primers. Four different primers two (forward and reverse) primers specific to SNP and two (forward and reverse) primers specific to non-SNP allele are used in the reaction.
The introduction of mismatched nucleotides at the 3’ primer end helps effective amplification of the SNP allele. After the amplification, the results are analyzed using a conventional agarose gel electrophoresis technique.
Depending on which set of primers amplify the allele, the SNP results have been validated. ARMS-PCR is widely used for the screening and diagnosis of beta-thalassemia and sickle cell anemia SNP mutations.
Advantages and Limitations of Allele-specific PCR:
| Advantages | Limitations |
| Simple and cost-effective | May require optimization for each SNP |
| High specificity and sensitivity | Limited multiplexing capability |
| Suitable for low-throughput applications | Susceptible to PCR biases and allelic dropout |
| Rapid results | Challenging for large-scale genotyping projects |
RFLP-based Genotyping:
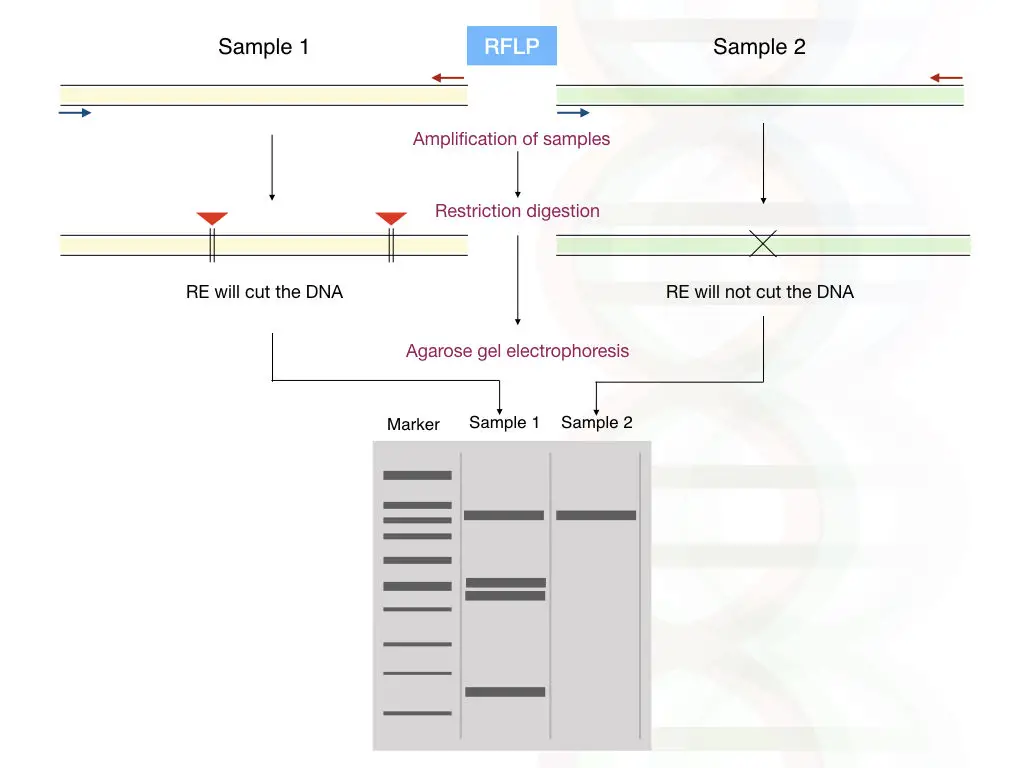
Another PCR-based genotyping method is RFLP. Refers to Restriction Fragment Length Polymorphism, RFLP is a combination of PCR amplification and restriction digestion by a specific endonuclease enzyme.
After the selective amplification of a DNA fragment containing the SNP, the allele is digested using a restriction enzyme. The restriction digestion protocol is designed in a way that the enzyme can either cut an SNP-containing allele or a normal allele.
The results are analyzed using an agarose gel electrophoresis technique. For instance, if the normal allele contains the restriction site, two or more DNA fragments (depending on the number of restriction sites) are observed in a gel for the normal allele.
The results would show two DNA bands for the normal allele, three for heterozygous and a single DNA band for the SNP allele. To learn about results and analysis you can read this guide: A Guide to Read and Understand Restriction Digestion Results.
Advantages and Limitations of PCR-RFLP:
| Advantages | Limitations |
| Simple and widely applicable | Requires prior knowledge of restriction enzyme sites |
| Cost-effective | Limited multiplexing capability |
| High specificity and sensitivity | Susceptible to PCR biases and allelic dropout |
| No specialized equipment required | Challenging for large-scale genotyping projects |
| Limited throughput |
TaqMan Probe-based SNP Genotyping:
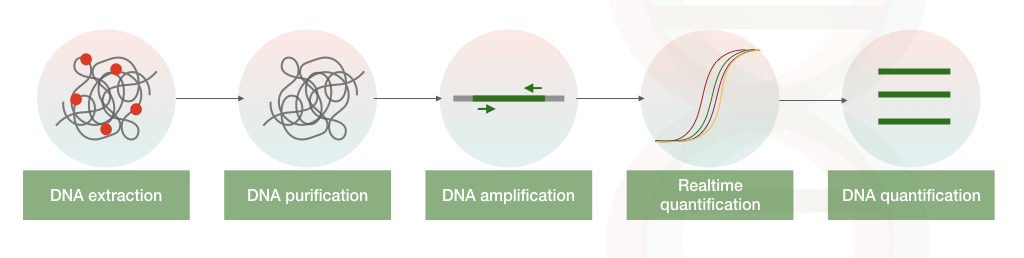
The present method is a quantitative PCR-based SNP genotyping technique. Here, a fluorescently labeled probe is used for the detection. Along with the set of primers, an SNP-specific TaqMan probe is used in the reaction.
The probe can only hybridize with the SNP allele. When the Taq DNA polymerase removes the probe during the amplification, the release of fluorescence gives the signal for positive amplification.
The probe can not bind with the normal allele. Measuring the fluorescence in real time determines the genotype of the sample. Notedly, the present method doesn’t need gel electrophoresis.
Advantages and Limitations of TaqMan Probe-based PCR Analysis:
| Advantages | Limitations |
| High specificity and sensitivity | Requires specific probes for each SNP |
| Real-time detection enables rapid analysis | Limited multiplexing capability |
| Quantitative measurement of allele abundance | Costlier than other PCR-based genotyping methods |
| Automatable and suitable for medium-throughput | Susceptible to PCR inhibition and non-specific amplification |
| Less prone to contamination compared to endpoint assays | Requires specialized equipment (qPCR) and software |
| No post-PCR processing required |
High-Resolution Melting Curve Analysis:
High-resolution melting curve analysis is yet another qPCR-based method used routinely for SNP genotyping. The melting temperature behavior varies from sequence to sequence, even a single base change in the sequence alters the melting temperature.
Using the melting property, the sequence is melted by gradually increasing the temperature. The use of double-stranded DNA dye, for instance- SYBR green dye, monitors the dissociation of various DNA fragments.
The presence of SNPs can alter the melting behavior, resulting in distinguishable melting curves for different genotypes. The present technique doesn’t need gel electrophoresis and is fast, cost-effective and accurate.
Advantages and Limitations of High-Resolution Melting Curve Analysis:
| Advantages | Limitations |
| Rapid and cost-effective SNP genotyping | Limited to known SNP sites |
| No post-PCR processing required | Sensitive to PCR artifacts and non-specific products |
| medium throughput and automation-compatible | Challenging for complex genotyping assays |
| Applicable for SNP discovery and screening | Sensitivity may vary depending on PCR conditions |
| Real-time analysis allows immediate results | Requires optimization for specific SNP assays |
Related article: TaqMan Probe vs SYBR green Dye-based qPCR- How to select the one?
KASP-based SNP genotyping:
KASP stands for Kompetitive Allele-Specific PCR and is a qPCR-based medium-throughput genotyping method. Much like the conventional AS-PCR, two separate forward primers along with a common reverse primer have been used for competitive amplification.
Each allele-specific primer is labeled with a different fluorochrome and allowed to amplify in a competitive amplification reaction. The primers anneal and amplify their respective allele and give the fluorescent signal.
In this way, the presence of an SNP allele is determined using an SNP-specific fluoro-labeled primer. The present method is more accurate and efficient and than other approaches in this category.
Advantages and Limitations of Kompetitive Allele-Specific PCR (KASP):
| Advantages | Limitations |
| Medium-throughput SNP genotyping | Initial setup costs may be higher |
| Accurate and reliable results | Requires optimization for specific SNP assays |
| Multiplexing capability for simultaneous analysis | Sensitivity may vary depending on SNP complexity |
| Cost-effective compared to other methods | Limited to known SNP sites |
| Applicable for various sample types and sizes | Complex genotyping assays may require validation |
Digital PCR:
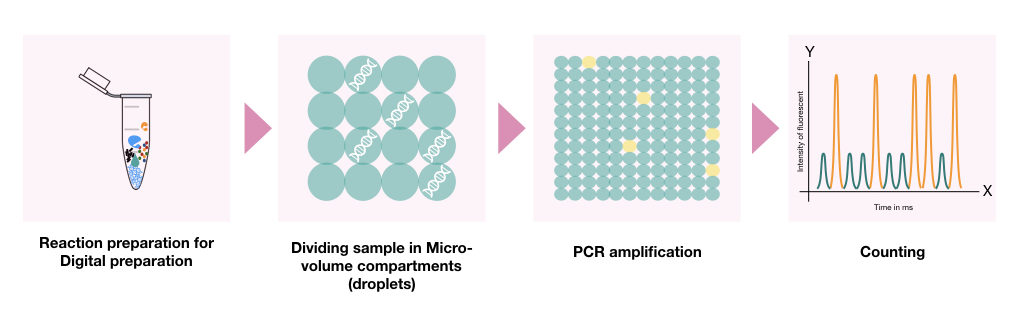
Digital PCR is an advanced and sensitive PCR approach for low-throughput SNP genotyping experiments. Here, the DNA sample is partitioned into thousands of separate reactions using oil-water emulsion droplets or microfluidics.
Each droplet contains a single or a few DNA molecules. Now, each separate reaction is performed using a set of primers and SNP-specific probes. The reaction or droplet containing our template allele can only be amplified and quantified.
Digital PCR increases the sensitivity and specificity of the genotyping and thus can perform genotyping for low abundance alleles.
Advantages and Limitations of dPCR for SNP Genotyping:
| Advantages | Limitations |
| Absolute quantification of target DNA molecules | Limited throughput compared to other methods |
| High precision and sensitivity for detecting rare variants | Costlier than traditional PCR and qPCR |
| Insensitive to PCR inhibitors and amplification biases | Requires specialized equipment and expertise |
| Enables accurate quantification of SNP allele frequencies | Sensitivity to sample quality and DNA integrity |
| Suitable for applications requiring precise quantification |
Microarray-based SNP Genotyping
The microarray-based genotyping technique is a hybridization-based method for the detection of not only SNPs but also indels, inversions and translocation. The present method is high-throughput, accurate and reliable, and can determine thousands of SNPs in a single run.
SNP Microarray Genotyping:
As aforementioned, SNP microarray is a high throughput and precise method for SNP genotyping that can study single base change from the entire genome. Here, the hybridization chip is filled with labeled DNA probes specific to various SNPs.
Now, the DNA sample is allowed to hybridize with the chip oligos, and hybridization emits fluorescence signals. The recorder records the signal and provides output as the presence or absence of SNP.
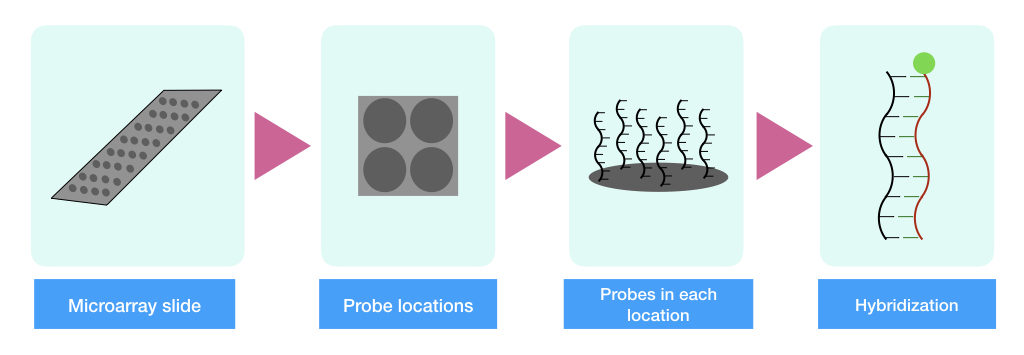
To know more about the whole process, I have given the link to the main article. You can read it there. Advantages and Limitations of SNP Microarray Genotyping:
| Advantages | Limitations |
| High-throughput SNP analysis | Requires prior knowledge of SNP locations |
| Simultaneous genotyping of thousands to millions of SNPs | Limited sensitivity for rare variants |
| Suitable for large-scale genetic studies | Data interpretation may be complex |
| Applicable for population-based studies | Costlier than PCR-based methods |
| Enables genome-wide association studies (GWAS) | Technical expertise required for data analysis |
Sequencing-based SNP genotyping
Sequencing techniques such as Sanger sequencing, targeted sequencing, pyrosequencing and NGS are powerful methods to genotype various SNPs. The present approach provides sequence-based SNP information. In addition, it can identify new SNPs present in the target sequence or from the genome.
Sanger Sequencing-based SNP genotyping:
The present method is based on the traditional Sanger sequencing technique using a ddNTPs-labeled chain termination approach. Here, the target region or a gene is amplified and sequenced in a sequencing machine.
Computational tools analyze the sequencing signals and prepare a chromatogram and DNA sequence. Using bioinformatics tools the sequence is compared and analyzed to the reference sequence.
This way, the SNP, as well as other mutations and new sequence alterations are characterized and studied.
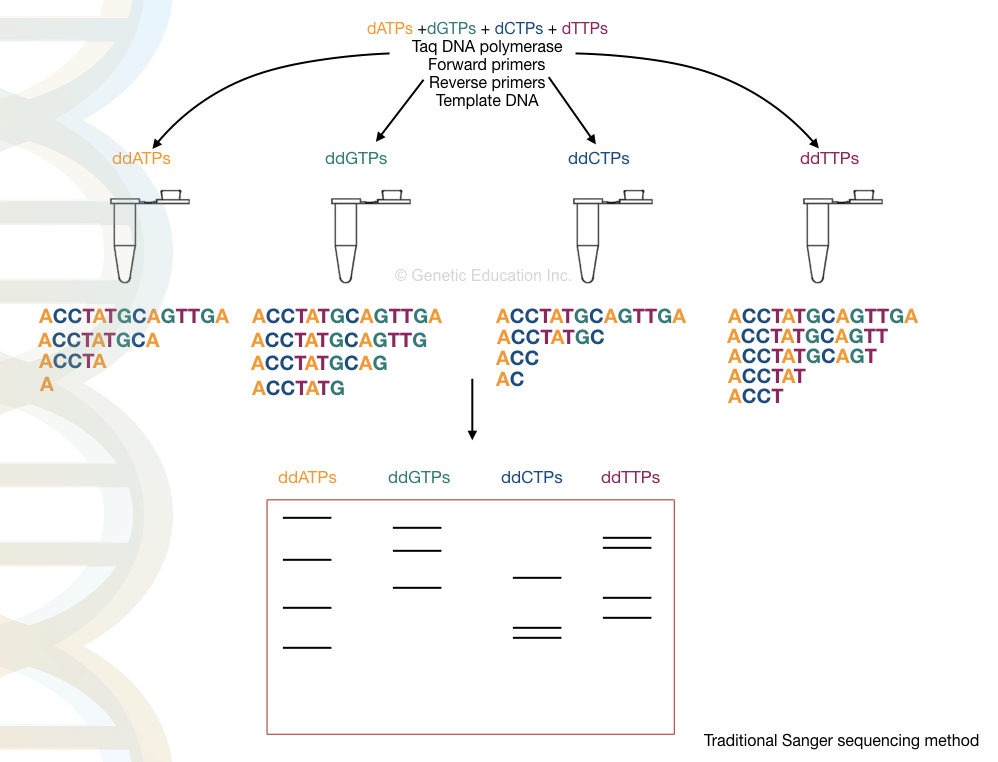
Advantages and Limitations of Sanger Sequencing-based SNP Genotyping:
| Advantages | Limitations |
| Highly accurate and reliable sequencing results | Relatively low throughput for large-scale studies |
| Suitable for validating SNP discovery findings | Limited scalability for high-throughput analysis |
| Precise determination of SNP allele | Longer turnaround time compared to other methods |
| Provides sequence information beyond SNP locus | Costlier than some high-throughput methods |
| Widely available and established technology |
Targeted SNP Genotyping:
Another sequencing approach widely used for SNP genotyping is targeted SNP genotyping. It’s although much similar to the Sanger sequencing, but with a small modification.
The sequencing run is designed around the SNP and not a gene or sequence. Meaning, that the sole purpose is to study the known, pathogenic and target SNP and not the sequence.
Amplicon-based sequencing, hybrid capture-based sequencing and molecular inversion probe (MIP) technology are several popular and high-throughput targeted SNP genotyping techniques.
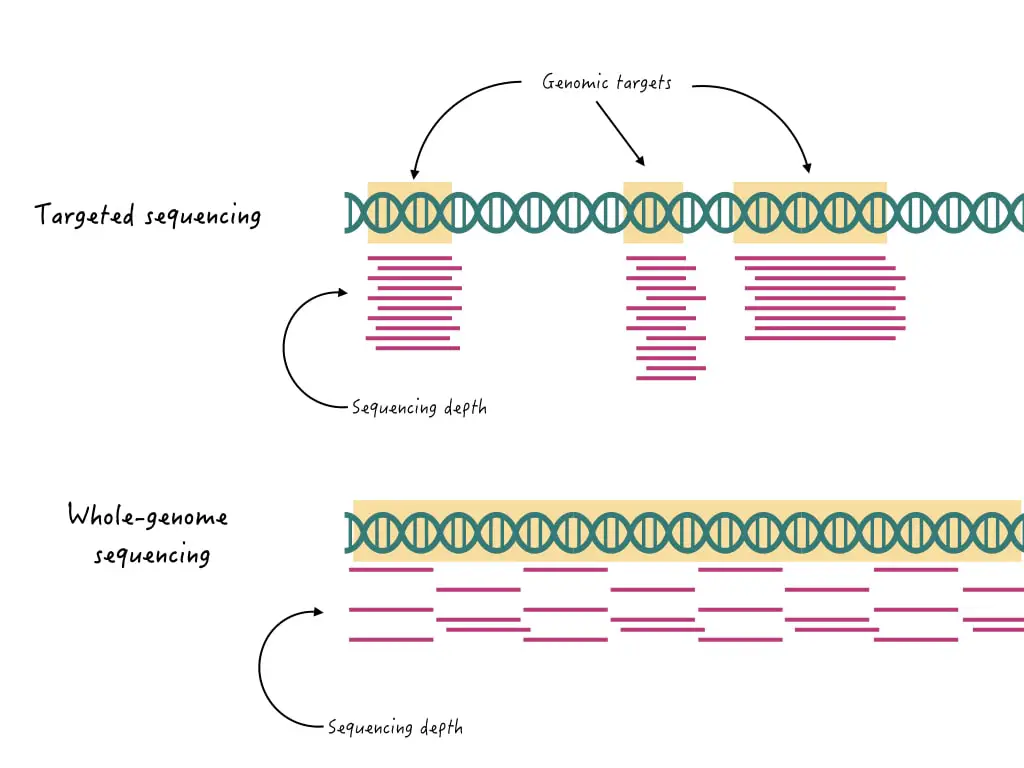
Advantages and Limitations of Targeted Sequencing for SNP Genotyping:
| Advantages | Limitations |
| Enables efficient and cost-effective SNP genotyping | Requires prior knowledge of SNP positions |
| Provides high specificity and coverage of targeted regions | May miss novel or rare SNPs outside the target regions |
| Allows for multiplexing of SNP assays in a single sequencing run | Initial design and optimization of targeted panels can be time-consuming |
| Suitable for studying specific genomic regions or gene panels | Sensitivity to sequence capture efficiency and uniformity |
| Enables customization of sequencing panels for specific research needs |
NGS-based SNP genotyping:
The present approach uses the power of next-generation sequencing technology to identify and genotype SNPs across a targeted genomic region or the entire genome. It’s an ultra-high throughput sequencing approach that can study millions of sequence alterations including the SNPs from the entire genome.
Here, DNA samples are first prepared and fragmented into smaller pieces. Adapters are then ligated to the DNA fragments, allowing them to be amplified and sequenced using a selected NGS platform.
Common and popular NGS platforms are Illumina, Ion Torrent, PacBio and Oxford Nanopore. During sequencing, each base in the DNA fragments is read multiple times, providing high coverage and accuracy for SNP calling.
Bioinformatics analysis is then performed to align the sequencing reads to a reference genome, identify SNPs, and determine their genotypes based on the sequence variations observed. Various software tools and algorithms are available for SNP calling, variant annotation, and downstream analysis of the genotyping data.
Advantages and Limitations of NGS-based SNP Genotyping:
| Advantages | Limitations |
| High-throughput genotyping of thousands to millions of SNPs | High initial setup and sequencing costs |
| Genome-wide coverage allows for comprehensive SNP analysis | Requires specialized bioinformatics expertise |
| Enables discovery of novel SNPs and rare variants | Data storage and management can be challenging |
| Suitable for population genetics studies and complex trait analysis | Sensitivity to sequencing errors and biases |
| Facilitates integration with other omics data for multi-omics analysis |
Related article: 7 Ways to Determine Genotypes Using Gel Electrophoresis.
How to select an SNP genotyping technique:
Now, so far we have discussed various techniques that we can use for genotyping, particularly for SNP genotyping. You may have a question, which technique is suitable and best for you?
Check out the table first.
| Technique | Principle | Advantages | Disadvantages | Throughput | SNP No. |
| PCR-Based SNP Genotyping | Amplification-based technique. Requires gel electrophoresis and fluorescent detection. | Simple and cost-effective | Limited multiplexing capacity for some methods | Low | 1 to 20 |
| Microarray-Based SNP Genotyping | Hybridization-based technique. | High throughput and multiplexing capacity | Limited sensitivity for rare variants | High | >1000 |
| Sequencing-Based SNP Genotyping | Direct or targeted sequencing of DNA regions containing SNP loci | High specificity and resolution | Higher cost and complexity than other methods | Moderate to ulta-high | A few 100 to millions |
- If you are a dissertation student and you want to study a single or a few SNPs, you can choose any PCR-based method. Ideally, RFLP is the best and most effective method for such experiments.
- However, other methods such as ARMS or qPCR can also be used, but it can increase the budget of the project.
- If you want to choose the technique for your project or PhD work you can combine PCR with DNA sequencing or simply you can use either a microarray or NGS-based approach for high-throughput analysis.
- If your experiment is large, and you want population-based, disease-based or genome-wide SNP data, you can use whole-genome next-generation sequencing or ultra-high throughput SNP array.
Wrapping up:
In conclusion, among many available SNP genotyping methods, the choice depends on your experimental requirement and the throughput you want. Nowadays, SNP genotyping methods such as ARMS-PCR, qPCR, and targeted sequencing high throughput NGS platforms are used in clinical diagnosis as well.
In addition, NGS and microarray techniques are the first choice in cancer research and related studies. I hope this article added value to your genetic learning. Do share this article and subscribe to our blog.
Resources:
Lawrie RD, Massey SE. Agrigenomic Diversity Unleashed: Current Single Nucleotide Polymorphism Genotyping Methods for the Agricultural Sciences. Applied Biosciences. 2023; 2(4):565-585. https://doi.org/10.3390/applbiosci2040036.
Perkel, J. SNP genotyping: six technologies that keyed a revolution. Nat Methods 5, 447–453 (2008). https://doi.org/10.1038/nmeth0508-447.
Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001 Sep 1;29(17):E88-8. doi: 10.1093/nar/29.17.e88. PMID: 11522844; PMCID: PMC55900.



