“Native gel electrophoresis helps in genetic screening and diagnosis by determining genotypes and identifying homozygous and heterozygous. This article explains 7 ways that this technique can be used for genotyping.”
Electrophoresis is a technique to separate molecules like DNA or RNA based on their size and electrical charge. This property allows us to visualize the DNA, do quality checks, validate PCR products and investigate genotypes.
However, native gel electrophoresis has been largely replaced by newer techniques such as capillary electrophoresis and real-time PCR for the diagnosis of genetic diseases. It can still be used in certain experiments. We have been using the present technique for genotyping, in our lab.
It worked and still works.
In particular, we were using it for prenatal diagnosis of thalassemia and sickle cell anemia. So how does it work? From my past experiences, I will explain 7 ways to use native gel electrophoresis for genotyping.
Stay tuned.
You should have knowledge of genotype to understand this article. Read this article to learn: What is a genotype?
Key Topics:
Determine Genotypes Using Gel Electrophoresis
Electrophoresis is extensively used in VNTR and STR genotyping. Experts analyze the banding patterns of various hypervariable loci and prepare the results. The present scheme is popularly used in forensic analysis and several other studies.
It also allows us to study single nucleotide polymorphisms (SNP), deletions, insertions and other alterations for a gene. Furthermore, homozygous and heterozygous conditions can be effectively ruled out for genotyping. Each technique is explained here.
SNP genotyping:
SNPs are alterations at a single nucleotide that certainly can’t alter the size of DNA and therefore technically we can’t investigate them directly through native gel electrophoresis. Certain optimizations, however, permit us to determine each genotype.
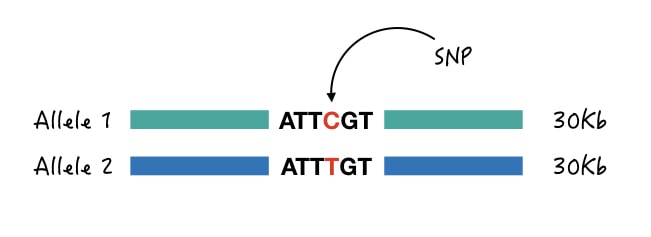
1. Size-difference genotyping:
Size differentiation is a highly unusual way for SNP genotyping but it works effectively. In this technique, ARMS primers having one or two mismatch bases are used for amplification.
Two separate forward primers are designed which targets each separate allele (wild and mutant allele) while two different reverse primers are designed in such a way that it creates size differences.
So in a gel, if the sample is normal, DNA bands will appear for homozygous wild type only, but not for homozygous mutants. And vice versa. However, as we have used two separate reverse primers to create size differences, two separate bands will appear in the heterozygous condition.
So, even though a single nucleotide is altered at the same position, we can determine genotypes by creating size differences in gel electrophoresis, in combination with the ARMS-PCR. The present approach is popularly used for thalassemia and sickle cell PCR.
This technique is impressive enough to distinguish homozygous and heterozygous.
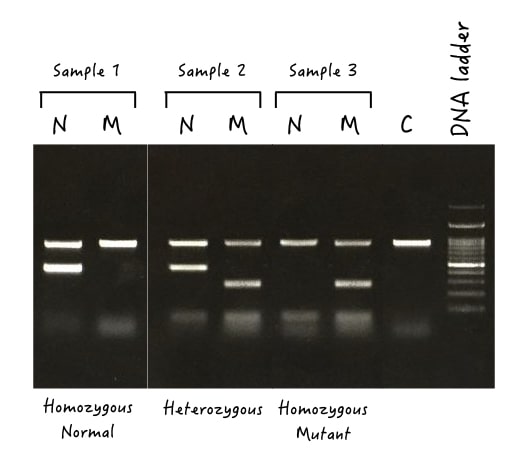
2. Genotyping of same-sized fragments:
PCR designing is a complex and tedious process. The success depends on how effectively the primers are designed. Oftentimes, it happens that to maintain efficiency we have to amplify each allele with the same set of primers. Meaning, it results in same-sized fragments.
Here, as the size difference can’t work, gel electrophoresis becomes a crucial detection factor. What we can do here is we run two different reactions for the sample, one with the normal set of primers and the other one with the mutant set of primers.
If both lanes show bands, the sample is heterozygous, if only a single lane shows the DNA band, the sample is either homozygous normal or wild type.
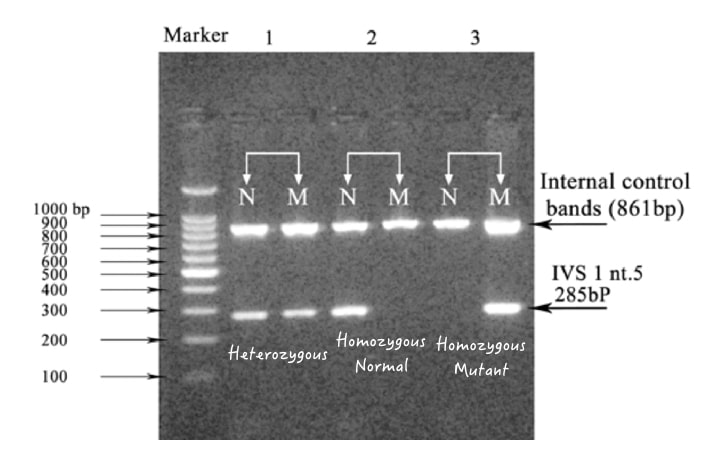
3. Single genotyping:
In this technique, only single allele-specific primers are added for amplifications, either for wild type or mutant type with the same trick explained above. And another allele can not be allowed to amplify.
Only a single prominent band will appear either in the homozygous wild type or in the homozygous mutant type. Single genotyping by gel electrophoresis has several crucial limitations. It can’t distinguish heterozygous alleles and we can’t determine non-specific amplification if it occurred.
Not advised for diagnosis of genetic diseases.
4. Genotyping for larger indels:
“Indels are insertions and deletions.”
Unlike SNP genotyping, larger deletions, insertions or duplications >100bp can be determined precisely using gel electrophoresis too. This is the most simple approach and requires a simple PCR setup.
A single set of forward and reverse primers are allowed to amplify the gene. Depending on, if any fragment is deleted, duplicated or inserted, it creates size differences in the gel and determines each genotype. For example, Huntigton’s disease.
Huntington’s disease occurs by genetic anticipation of CAG triplet repeat in which the expansion of the present triplet results in the disease state. So when we use a single set of PCR primers, we can amplify the homozygous normal, mutant or heterozygous alleles present in the sample.
Each allele separates depending on the size (addition of the CAG repeats) of the gene, thereby determining each genotype. Scientists use the present approach popularly for the screening of triplet repeat disorders, for instance, Fragile X syndrome, Myotonic dystrophy and spinocerebellar ataxia.
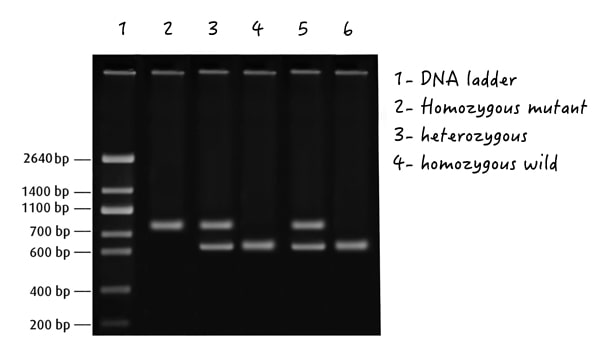
5. Genotyping by restriction digestion gel electrophoresis
Yet another common and classic technique for ‘genotyping by gel electrophoresis’ is restriction digestion. In this, a particular restriction enzyme cleaves the amplified gene, and when run on a gel, separates alleles.
The restriction enzyme is selected in a way that it either cuts the wild type or mutant type allele, but not both. The digested, undigested and heterozygous products show two, single and three or more separate bands in a gel.
Note:
Restriction digestion is the best technique to discriminate homozygous (normal and mutant) and heterozygous alleles, as it has higher banding resolutions. Note that restriction digestion is performed on 3% gel.
image
6. STR genotyping:
The native gel electrophoresis is also capable enough to do STR genotyping. STRs are short tandem repeats present in our genotype and are highly variable in size. Different alleles have a different number of repeats which gives us the power to discriminate alleles.
It is used in DNA testing, paternity, maternity and parental DNA testing, criminal verification and finding blood relations.
7. VNTR genotyping:
Yet another popular marker for DNA testing is VNTR which is a variable number of tandem repeats present one after another in the genome and are highly variable. VNTR genotyping follows the same process and steps as the STR but as they are larger in size give higher gel resolution.
How to determine homozygous vs heterozygous in a gel?
So far, we have discussed ways to determine genotypes using the native gel, but you may wonder how we can determine homozygous vs heterozygous in a gel. It’s quite a difficult task, actually. Starting with the definition:
Homozygous is a condition of having two identical alleles, either both wild type or both mutant type whereas heterozygous is a condition of having two different alleles, one wild and another mutant type allele. So how can you distinguish them? See the image first.
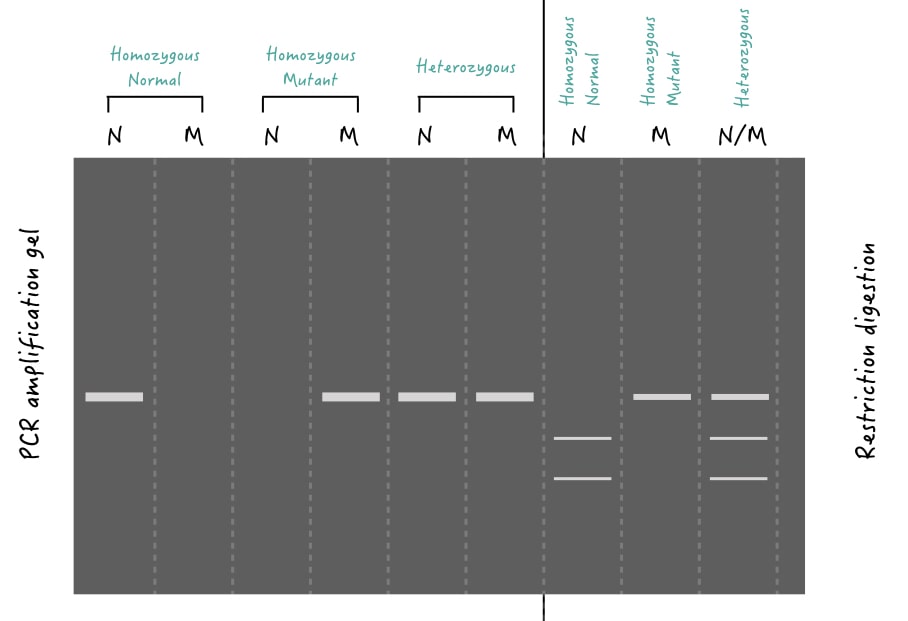
In the case of homozygous, both identical chromosomes comprise the same allele (same in size, nucleotide sequence and location). So if we run a PCR gel, only a single dominant band will be observed (Because both are the same!). However, in restriction digestion, depending on the presence or absence of a restriction site, two/more or single bands will appear, respectively.
See lane 1 for homozygous normal, lane 3 for homozygous mutant and lanes 7, 8 and 9 for restriction digestion.
In the case of heterozygous, both identical chromosomes compare different alleles (different in either size or sequence) and hence if we run a PCR, two different primer sets amplify each allele thereby giving two different DNA bands in the gel. For restriction digestion, depending on how many restriction sites the target allele has, 3 or more DNA bands will appear.
See lanes 5 and 6 for heterozygous alleles detected by PCR and lane 9 for heterozygous alleles detected by restriction digestion.
Read more: Genotype vs phenotype.
Wrapping up:
In conclusion, gel electrophoresis is capable enough to perform genotyping, and people still use it even in prenatal screening. It’s a powerful tool that can separate each allele based on their size differences.