“Metagenomic analysis has revealed crucial information regarding microbial diversity. Explore the hidden world of microbes by joining this fascinating journey. Here are 10 amazing facts revealed by Metagenomics.”
Have you ever wondered about the hidden microbial world around us? How complex and diverse it is! Do you know what we know about microbes is the tiniest part of it and the huge portion is still unknown to us!
Microbes have significant importance for life on earth to survive and sustain our ecosystem. They are a crucial link in our ecosystem and are present, from the soil to the air we breathe. That’s until the recent advancement and research in metagenomics, we certainly know about it.
Metagenomics is an interdisciplinary field of genomics and microbiology that allows scientists to study a vast diversity of microbes present in any ecosystem or host. Cutting-edge techniques like NGS and shotgun sequencing make it possible for us.
We can explore the diversity and unknown portion of this unexplored living mass through metagenomics. In this article, I am listing amazing facts revealed by various metagenomic research which will blow you away.
At Genetic Education Inc. our motto is to serve knowledge. We have researched the web, read tons of articles and extracted this amazing information for you. Hope you will like it.
Stay tuned.
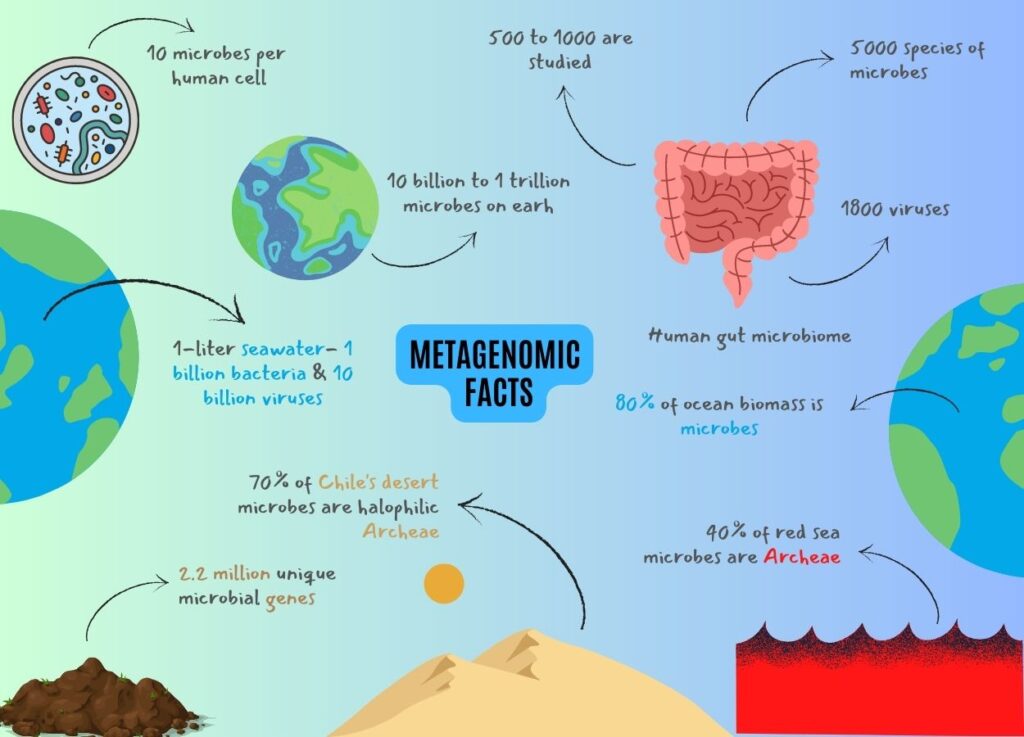
Mind-Blowing Facts Revealed By Metagenomics
- The microbial diversity of planet Earth is immense. 10 billion to 1 trillion microbial species are present on Earth in different regions, atmospheres and environments; and fascinatingly, only a small part is scientifically well-characterized and known.
The human gut microbiome is a complex network of varied microbes having a significant role in health and well-being. Thus, it has been a key research focus for years. Over 5000 peer-reviewed articles on the topic of “gut microbiome metagenomics” are present only on PubMed. Imagine how important the topic is.
- A study published in the Nature Journal in 2012 reported 3.3 million unique genes from the gut samples of 124 European individuals. Interestingly, our human genome contains only 20,000 genes, roughly. Imagine how diverse only the profile of the gut microbiome is!
- The human microbiome project was launched in 2010 to study the microbial community in and around humans using metagenomic techniques. The project has sequenced over 5000 samples from 250 healthy individuals, and identified, very first time, 5000 different microbial species from the samples.
- In estimation, scientists have identified and classified 500 to 1000 gut microbe species using metagenomic analysis, till now.
- One fact also suggests that the gut microbiome profile has a key role in obesity and metabolic disorders as the gut microbe of an obese individual is significantly different from that of a healthy individual. This shows that the gut microbiome has a pivotal role in causing lifestyle diseases and other health conditions.
- However, despite the present fact, a study published in Cell (2019), suggests that the core microbial taxa are common among all individuals from different geographical locations. The study included 6 continents, 747 individuals and 9,428 samples.
Thus, factors like diet, geographical location, age, genetics and environment of individual contributions to the gun microbiome diversity.
- In addition to just microbes, the metagenomic analysis further adds more value to viral research and helps understand the complex diversity of viruses. In 2019, a study published in Science Journal demonstrated that over 1,800 different viruses are found in the human gut profile. Among them, the majority are still unknown to us and can infect us.
- One interesting piece of information also shows that we are more microbes than humans! As our body contains 10 times more microbial cells than our own cells. Isn’t it shocking!
Microbes also have an important role in the environment and different ecosystems as they can break down organic components, convert molecules from one to another form, perform nitrogen fixation, produce secondary metabolites, etc.
- A metagenomic study revealed that the global soil microbiome contains over 2.2 million unique genes which give incredible diversity to different soil ecosystems across the globe (A study published in Science in 2017 on different soil samples collected across various religions of the world).
Metagenomic studies also revealed amazing data regarding antibiotic resistance as well. Antibiotic or antimicrobial resistance is a well-established phenomenon in which microbes develop resistance against antibiotics and become competent for survival. Interestingly, antibiotic resistance is prevalent in the microbes present in various sources like water, air, soil and mud (wastewater), etc.
- They develop such genes to compete for resources and survival. One study published in the Journal of Global Antimicrobial Resistance in 202 reported that over 60% of study samples collected from various sources contain one or more antibiotic resistance genes.
Interestingly, such valuable information helps researchers to develop new strategies and antibiotics against various pathogens and microbes.
Metagenomic studies also shed light on the third domain of life ‘archaea’ and give us lucrative knowledge regarding how amazing archaea is. Studies depict that archaea are distinct from eukaryotes and prokaryotes and can live in diverse environments and extreme habitats like permafrost, deep-sea vents, hot springs or even at extreme temperatures.
- A study published in 2019 on the metagenomic analysis of the red sea, showed that from the total microbial diversity, archaea account for 40% of the red sea microbial community. Interestingly, the major archaea detected was novel.
- As aforesaid, archaea are ‘extreme environment-loving’ organisms. Another study revealed that archaea are the dominant microbial community present in the Atacama Desert Chile. As the region is highly salt concentrated, 70% of total archaea reported were halophilic- salt-loving.
- Microbes also have a crucial role in the ocean ecosystem and are hugely diverse, much like other microbial populations. It is estimated that 1 liter of seawater contains 1 billion bacteria cells and 10 billion viruses, roughly.
- In addition, 98% of total oceanic biomass is just single-celled microbes! Including bacteria, viruses, protozoa, plankton, fungi and archaea.
- Previously it was believed that the woman’s placenta might be highly sterile, however, metagenomic studies revealed that the placenta sample contains a diverse group of microbes and falsify the assumption.
Interestingly, it’s comparatively a tiny population, but entirely different from the maternal and paternal microbiome. Scientists believe that the placenta microbe should have a defined role in the pregnancy’s progress.
Wrapping up:
Metagenomic analysis revealed a wealth of data, information and knowledge regarding microbes around us. Such data and studies collectively help scientists to understand the complex nature of microbes, their diversity and their metabolic activity thereby providing ways to develop new strategies to improve our health and well-being.
In addition, metagenomic studies will also assist in improving various ecosystems. Collectively, the possibilities are endless from personalized medicine to pharmacogenetics. And preserving ecosystems to produce byproducts like bio flue or synthetic nutrients.
I hope you like this amazing information and data on microbes. I advise, if you are interested in this topic, please read our previous article on the present topic. Those help you to learn metagenomics more precisely.
Read next:
- Metagenomics Made Easy: Streamlining DNA Extraction With Bead Beating.
- A Guide To Next-Generation Shotgun Sequencing In Metagenomics: Technique, Advantages and Challenges.
Useful resources:
- The Human Microbiome Project website: https://hmpdacc.org/
- The paper “Structure, function and Diversity of the healthy human microbiome” published in Nature: https://www.nature.com/articles/nature11234
- The paper “A global atlas of the dominant bacteria found in soil” published in Science: https://science.sciencemag.org/content/359/6373/320
- The paper “The global prevalence and distribution of antibiotic resistance in natural environments: a systematic review and meta-analysis” published in the Journal of Global Antimicrobial Resistance: https://www.sciencedirect.com/science/article/pii/S2213716520301622
- The report “Metagenomics Market by Product, Technology, Application, End User – Global Forecast to 2025” by Research and Markets: https://www.researchandmarkets.com/reports/5048202/metagenomics-market-by-product-technology
- The paper “An atlas of human microbiome composition four years in the making” published in Science: https://www.sciencemag.org/news/2019/04/atlas-human-microbiome-composition-four-years-making
- The paper “The placenta harbors a unique microbiome” published in Nature Communications: https://www.nature.com/articles/s41467-018-08901-3
- The paper “Geography and location are the primary drivers of office microbiome composition” published in PLoS One: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0132788
- The Human Microbiome Project Consortium. (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486(7402), 207–214. https://doi.org/10.1038/nature11234
- The Human Microbiome Project Consortium. (2012). A framework for human microbiome research. Nature, 486(7402), 215–221. https://doi.org/10.1038/nature11209
- Franzosa, E. A., Hsu, T., Sirota-Madi, A., Shafquat, A., Abu-Ali, G., Morgan, X. C., Huttenhower, C. (2019). Sequencing and beyond: Integrating molecular ‘omics’ for microbial community profiling. Nature Reviews Microbiology, 17(11), 731-744. https://doi.org/10.1038/s41579-019-0244-4
- Lloyd-Price, J., Abu-Ali, G., & Huttenhower, C. (2016). The healthy human microbiome. Genome Medicine, 8(1), 51. https://doi.org/10.1186/s13073-016-0307-y
- Human Microbiome Project Consortium. (2012). A framework for human microbiome research. Nature, 486(7402), 215–221. https://doi.org/10.1038/nature11209
- Pasolli, E., Truong, D. T., Malik, F., Waldron, L., Segata, N. (2016). Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Computational Biology, 12(7), e1004977. https://doi.org/10.1371/journal.pcbi.1004977

