“Want to master Sanger sequencing? Explore this article to learn the advantages and limitations of Sanger sequencing with technical specifications.”
Sanger sequencing is a gold-standard diagnostic technique, relying on the chain termination chemistry. Automations, recent advancements in sample processing and highly sophisticated analysis software are crucial advantages, helping it to gain popularity.
In addition, there are several other reasons, scientists are still choosing it over the NGS! However, like all other technologies, Sanger sequencing comes up with limitations too, and that can’t be overlooked.
Want to know why Sanger sequencing is still popular and where it falls short?
In this article, we will deep dive into the advantages and limitations of Sanger sequencing. This article justifies why life science students should learn Sanger sequencing.
Stay tuned.
Key Topics:
Sanger sequencing advantages:
Sanger sequencing is a superior, popular and widespread technology for sequencing genes and DNA fragments. A few good advantages are discussed here.
Accuracy:
Sanger sequencing is extremely accurate with a ~99.999% correct base call rate. The reason being most accurate, is timely advancements.
Instrumental automation, kit-based approaches for extraction and sample preparation, well-developed and standard protocols and optimized analysis software are key factors in providing extreme accuracy.
When performed properly, it produces clear, accurate and high-quality reads with minimal background noise in Sanger sequencing.
However, it struggles with sequencing repetitive, homopolymeric and high GC-rich regions, and where the accuracy is dramatically compromised.
Long reads:
One of the key advantages of Sanger sequencing is to produce long, continuous reads– typically around 600 to 100 bp. It is much better than the NGS (Next Generation Sequencing), having a short read length between 150 to 300 bp.
Long read length is an ideal and reliable setup for smaller genes and fragments, therefore, it is widely used for plasmid sequencing, gene cloning and validation testing and is also a primary choice for research and diagnosis.
On a technical side, it can accurately read 600 to 800 bps only with good preparations. Increasing read length usually compromises the accuracy. However, the read length is significantly shorter than the new age and third-generation real-time single-molecule sequencing.
Low sequencing errors:
Sanger sequencing is renowned for its exceptionally low error rate, typically as low as 0.001%. This precision comes from its unique chain termination chemistry, use of four different fluorescent dyes, and sophisticated capillary system.
The low error rate is the key factor that has been used for the detection of single-nucleotide polymorphisms. Even at low signal strength, optimized and traited analysis software provides exceptionally accurate results.
It is noteworthy that the accuracy is far better than the new-age NGS.
Compatible with low-quality and quantity DNA:
Sanger can easily deal with low-quality and quantity DNA and still yield amazing results. Again, because of the innovative chemistry, novel assays, kits and protocols and automation help achieve nearly perfect results.
Therefore, it is widely used for forensics, ancient DNA studies and rare variant analysis with low-quality and quantity DNA.
High-quality sequencing data:
One of the strongest advantages of Sanger sequencing is its ability to provide high-quality data. It generates sharp, distinct, well-spaced and clear chromatogram (sequencing) peaks.
The data can also be cross-validated by looking at both the chromatogram and the plain sequencing text file.
In addition, software is now powerful enough to understand the signals and prepare extremely precise base sequencing information even with ‘not-so-good’ sample preparation.
However, it struggles with the overlap, heterozygous or infection, background noise and other sequencing artifacts. Note that the quality highly depends on the sample preparation, primer design, reaction preparation and other technical factors.
Easy interpretation:
Four different colors for four different DNA bases! It’s that easy to interpret the results.
The output is typically a clear and distinct chromatogram where each colored peak corresponds to each nucleotide, making the analysis straightforward. The chemistry uses four different fluorescent-labeled ddNTPs for chain termination, providing four different separate color signals.
Even beginners can understand and interpret the results by looking at the electropherogram. No extensive bioinformatics skills or training needed.
No advanced bioinformatics needed:
Unlike NGS, which needed extensive bioinformatics tools, pipelines, programs and trained manpower, and highly enhanced computer and storage systems, Sanger relies on software-based analysis, just like our mobile application or computer software.
This makes it easy to use and understand and a bioinformatics minimal analysis.
Suitable for complex variants:
NGS leads on many fronts! For instance, it can’t accurately address complex variants like the tiny insertion or deletion or point mutation of several bases. Sanger does a better job in this scenario!
By strategising the complex variants, it can easily be investigated visually using Sanger sequencing. And therefore, it’s been employed for NGS data validation as well.
Cost-effectiveness:
Sanger sequencing is a super-cheap technique, in comparison with other available sequencing platforms. Its per-base sequencing cost is way lower than NGS. Cost is the major reason why it has been extensively used in diagnosis.
Ease in fragment analysis:
In the era of high-throughput sequencing, Sanger is still the primary choice for fragment analysis. Fragment analysis is a technique to determine DNA fragment sizes for genotyping, DNA testing and genetic analysis.
The present technique provides a single-base resolution between fragments with ~99.999% accuracy. Moreover, software-based fragment analysis aids in better results and analysis.
Automation and advancement:
Timely automations and advancements in Sanger sequencing make it a superior sequencing technology and still relevant even after 45 years. The two-touch automation, a latest innovation, allows the least human interference and high sequencing precision.
The availability of ready-to-kit reagents, assays for various mutations, genes and experiments, customized parts, and open-ended and free software makes it more valuable in recent times.
Simple workflow:
DNA extraction, amplification, sequencing and analysis, that’s it.
Sanger sequencing has an easy and simple workflow. It doesn’t have complex steps like library preparation, library enrichment, adaptor ligations and indexing. The simplest workflow makes it easy for even beginners to handle it.
No extensive computation work or kits are needed for sample preparation.
Limitations of Sanger sequencing:
Despite its widespread popularity, applications and utility, it still has several major drawbacks. A few are explained here.
Low throughput:
Sanger sequencing is a very low-throughput technique that enables sequencing only a few hundred nucleotides. Low throughput is the biggest disadvantage of the present approach.
Inefficient for WGS:
Due to this reason (low throughput), it isn’t ideal for whole-genome sequencing. It took more than 10 years for scientists to sequence a complete human genome using the Sanger sequencing method.
For larger-scale and whole-genome sequencing, NGS is ideally recommended.
Expensive for large-scale projects:
Extensive preparation and sample processing, and low throughput collectively limit its applications for larger-scale projects like WGS, WES or WMS due to high processing cost.
Sanger is a cost-effective technique for smaller projects, but expensive for large-scale projects and high-throughput analysis.
Inefficient for GC-rich, homopolymeric and repetitive regions:
These three regions of our genome are the most difficult to sequence, even with NGS. Sanger sequencing incorporates unwanted errors, background signals and wrong bases when it encounters GC-rich, homopolymeric and repetitive sequences.
Limited detection range:
It has a limited detection range due to low throughput, limited read length and difficulties studying rare variants. It faces challenges identifying very complex conditions like inversions, deletions and extremely rare variants.
Low sequencing depth:
Sanger sequencing has a very low sequencing depth, a base is sequenced once! Contrary, NGS has very high sequencing depth from 50 to 90X. Meaning each base has been sequenced 50 to 90 times during the parallel sequencing.
Low sequencing depth can’t give enough strength or confidence to report low-frequency variants.
Extensive assay design:
The present sequencing approach is easy, fast and accurate, but at the same time it needs extensive assay design. For example, for a 10 Kb gene, 12 to 15 different sequencing reactions are prepared.
This also needed extensive primer design, amplification steps and analysis.
Inefficient for detecting low-frequency variants:
Minor variants that exist in 15 to 20% of cell populations are often masked by the dominant allele, leading to false-positive or false-negative results.
Due to limitations of chemistry, sequencing depth, and detection range, the present technique is not suitable for low-frequency and rare variant studies.
Summary:
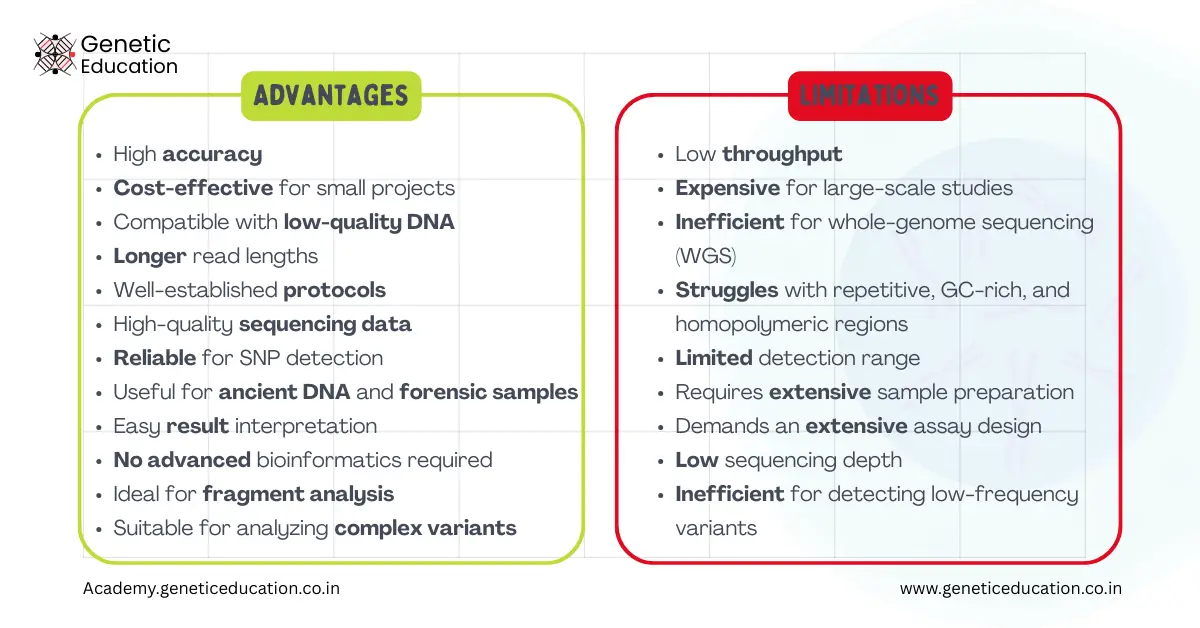
| Advantages | Limitations |
| High accuracy | Low throughput |
| Cost-effective for small projects | Expensive for large-scale studies |
| Compatible with low-quality DNA | Inefficient for whole-genome sequencing (WGS) |
| Longer read lengths | Struggles with repetitive, GC-rich, and homopolymeric regions |
| Well-established protocols | Limited detection range |
| High-quality sequencing data | Requires extensive sample preparation |
| Reliable for SNP detection | Demands an extensive assay design |
| Useful for ancient DNA and forensic samples | Low sequencing depth |
| Easy result interpretation | Inefficient for detecting low-frequency variants |
| No advanced bioinformatics required | – |
| Ideal for fragment analysis | – |
| Suitable for analyzing complex variants | – |
Wrapping up:
Sanger sequencing is, undoubtedly, unmatched and irreplaceable. It has a very high diagnostic value. In addition, the availability of ready assays, reagents and kits improved the TAT and speed of testing.
If you want to use Sanger sequencing for your experiment, these points will help you to make a decision. I hope you like this article. Subscribe to Genetic Education.
Resources:
- Manyana, S., Gounder, L., Pillay, M., Manasa, J., Naidoo, K. and Chimukangara, B. (2021). HIV-1 Drug Resistance Genotyping in Resource Limited Settings: Current and Future Perspectives in Sequencing Technologies. Viruses, [online] 13(6), p.1125. doi:https://doi.org/10.3390/v13061125.
- Baqur, M. and Hashim, H.O. (2023). Mastering DNA chromatogram analysis in Sanger sequencing for reliable clinical analysis. Journal of Genetic Engineering and Biotechnology, 21(1). doi:https://doi.org/10.1186/s43141-023-00587-6.


