“ddNTPs are known as chain termination nucleotides. In this article, learn about what ddNTPs are, why we are using them, and the difference between ddNTP and dNTP.”
PCR and DNA sequencing are two important and gold-standard methods in genetic research. dNTPs are one essential and common ingredient in Both. However, sequencing needs an additional ingredient that is ddNTP for chain termination.
dNTPs are known to us. We are using it in the PCR. Interestingly, the normal replication process also uses dNTPs to synthesize new DNA strands. Meaning that any DNA synthesis experiment needs deoxynucleotide triphosphates (dNTPs).
ddNTPs are entirely different from dNTPs. Not in synthesis, but ddNTPs are used for chain termination. Want to know how?
In this article, I will explain to you the concept of ddNTPs and their role in Sanger sequencing. I will also explain the differences between ddNTPs and dNTPs.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What are ddNTPs?
Massive advancements have been made in DNA sequencing within the last 25 years. Sanger and NGS sequencing platforms are considered a gold standard method for research and diagnosis.
Sanger sequencing is a powerful discovery in the field of genetics. It can sequence the DNA fragment using synthesis and chain termination. Much like the PCR, here also, the dNTPs have been used for synthesis, while specialized ddNTPs have been used for chain termination.
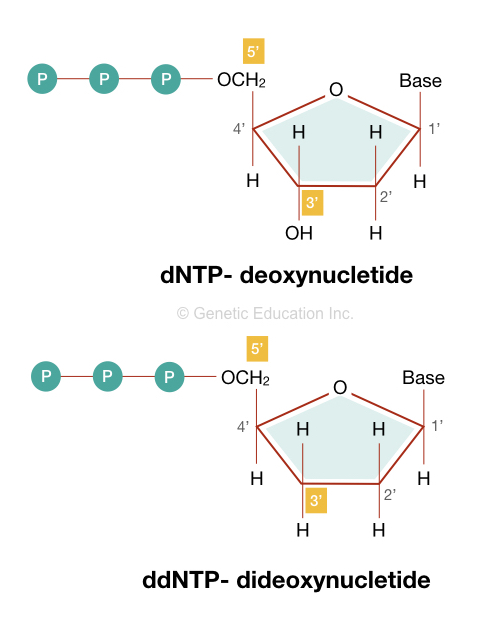
ddNTPs stand for dideoxynucleotide triphosphates. They are artificially synthesized and chemically altered forms of normal dNTPs that lack the 3’ OH (hydroxyl) group. The first reported evidence of the use of ddNTPs was in Sanger sequencing. Since then, it’s been used in the Sanger sequencing method.
ddNTPs are structurally similar to the dNTPs but lack the 3’ OH group on the sugar moiety of the nucleotide. The absence of a hydroxyl group prevents chain elongation as polymerase essentially requires it.
Structurally, it contains a pentose sugar which is a five carbon ring. Here a phosphate group is attached to the 5’ end while one nucleotide base (adenosine, cytosine, guanine and thymine) is attached to the 1’ carbon of the pentose sugar.
However, instead of the hydroxyl group at the 3’ end, ddNTP contains a single hydrogen atom. This will make it unique in comparison to the dNTPs.
Four different ddNTPs we use a sequencing reaction are ddATP (deoxyadenosine triphosphate), ddGTP (Deoxyguanosine triphosphate), ddTTP (Dideoxythymidine triphosphate) and ddCTP (Dideoxycytidine triphosphate)
Now, let’s see how it works.
How does it work?
3’ end of the nucleotide (dNTP) works as an initiation site for DNA synthesis in vivo (replication) as well as in vitro (DNA amplification). The DNA polymerase is only able to add complementary nucleotides if it can find a 3’ hydroxyl group on the pentose sugar.
The DNA polymerase settles there, uses the hydroxyl group and forms a phosphodiester bond. However, the entire system can not work if the DNA polymerase can not find the 3’ hydroxyl group.
Lack of 3’ hydroxyl group in ddNTPs results in termination of chain elongation.
How does ddNTP work in Sanger sequencing?
Now, it’s important to understand how this concept is beneficial in the Sanger sequencing method.
Sanger sequencing works on the principle of DNA synthesis and chain termination. DNA polymerase synthesizes the complementary DNA sequencing using the 3’ OH end of the dNTPs, however, it terminates the elongation when it encounters the ddNTPs.
Four different chain termination reactions occur for four different ddNTPs and resultantly produce different-sized DNA fragments. Importantly, the presence of a fluorescent tag on the ddNTPs helps determine the nucleotide at the termination site.
Using either a PAGE or capillary gel electrophoresis approach, various-sized fragments are analyzed to determine the order of nucleotides present in the sequence.
By labeling the ddNTPs with different fluorescent dyes, each terminating nucleotide can be identified, enabling the sequencing of the template DNA strand.
Related article: DNA Probes: Labeling, Types, Applications and Limitations.
Concentration
The ideal concentration of each ddNTP in the sequencing reaction is between 0.1 to 0.2 mM, however, the exact concentration depends on experimental requirements. It’s also noted that the ddNTP concentration should be kept lower than the dNTP concentration for effective sequencing.
dNTP to ddNTP ratio in the sequencing reaction should be 1:10 or more. For instance, if we add 0.1 mM ddATP we have to add 1 mM dATP. The concentration of ddNTP is intentionally kept lower for controlled termination.
By keeping the concentration of ddNTPs low, the probability of their incorporation at any given nucleotide position is minimized. This ensures that most of the DNA strands undergo elongation with regular dNTPs, allowing for the generation of DNA fragments of varying lengths terminated at different positions corresponding to the incorporated ddNTPs.
This differential termination pattern is essential for determining the sequence of the original DNA template during the sequencing process.
Differences Between ddNTPs and dNTPs:
- ddNTP stands for dideoxynucleotide triphosphate while dNTP stands for deoxynucleotide triphosphate.
- ddNTPs lack the 3’ hydroxyl (OH) group, on the other hand, dNTPs contain the hydroxyl group (OH) at the 3’ end.
- ddNTP contains a hydrogen atom at position 2’ and 3’ of the pentose sugar while the dNTP contains a hydrogen atom at only 2’ positive of the pentose sugar.
- ddNTPs terminate chain elongation and stop DNA synthesis. Conversely, dNTPs induce DNA synthesis by incorporating into a growing DNA strand.
- ddNTPs do not work as a substrate for DNA polymerase and stop the synthesis process while dNTPs work as a substrate for DNA polymerase and continue the synthesis process.
- Resultantly, ddNTPs can not form phosphodiester bonds while dNTPs can form phosphodiester bonds.
- ddNTPs are used only in DNA sequencing. On the other hand, dNTPs are used in DNA sequencing and PCR.
- In Sanger sequencing, ddNTPs are labeled with different fluorochromes. dNTPs do not contain any labels.
Related article: A Guide to Choose DNA Polymerase for Your Experiment.
Summary:
| Characteristic | ddNTPs | dNTPs |
| Full Name | Dideoxynucleotide triphosphates | Deoxynucleotide triphosphates |
| Structure | Lack of a 3′ hydroxyl group (OH) | Have a 3′ hydroxyl group (OH) |
| Hydrogen ions are present on both 2’ and 3’ carbon | 3’ carbon does not contain hydrogen ion | |
| Function | Terminate DNA strand elongation | Facilitate DNA strand elongation |
| Phosphodiester bond | Can not add phosphodiester bonds | Can add phosphodiester bonds. |
| Substrate | Does not work as a substrate for DNA polymerase | Work as a substrate for DNA polymerase |
| Presence in Sequencing | Used in Sanger sequencing for termination | Used as substrates for DNA synthesis |
| Termination Points | Generate termination points at specific bases | No specific termination points |
| Fluorescent Labeling | Labeled with different fluorescent dyes | Not fluorescently labeled |
| Role in Sequencing | Essential for determining the DNA sequence | Not involved in DNA sequence determination |
| Concentration in the sequencing reaction | Present at a low percentage (~0.1-0.2 mM) | Present at a higher concentration (>10 folds to ddNTPs) |
| Application | Used in DNA sequencing only | Used in DNA sequencing and PCR |
Read more on Sanger Sequencing:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- Detailed History of DNA Sequencing
- Why Should Life Science Students Learn Sanger Sequencing? – Top 6 Reasons
Wrapping up:
In conclusion, ddNTPs along with dNTPs play a vital role in DNA sequencing. Where dNTPs are involved in chain elongation, the presence of fluoro-labeled ddNTPs help in sequence determination.
I hope this article will clarify your doubts regarding ddNTPs and dNTPs. If you like this article, please share and subscribe to our blog.
Sources:
Slatko BE, Albright LM, Tabor S, Ju J. DNA sequencing by the dideoxy method. Curr Protoc Mol Biol. 2001 May;Chapter 7:Unit7.4A. doi: 10.1002/0471142727.mb0704as47. PMID: 18265267.