Do you know a typical NGS machine can generate around 1.8 to 6TB of sequencing data in a single run?
PCR duplication is a serious problem in so many crucial techniques like NGS, single-cell genomics, rare variants detection and quantification by digital PCR. PCR duplication means over-amplification of a single or certain type of template.
For example, in NGS (Next-Generation Sequencing), we can amplify the entire genome; over-amplification during library preparation can produce inefficient, inaccurate or biased results. Every fragment that we are going to add to the library must be amplified almost evenly.
Thus, conventional PCR can’t perform library enrichment for NGS, effectively. In addition, preparing a multiplex PCR reaction or setting up different PCR for every fragment from the library is also not possible, realistically.
So what scientists can do?
Scientists have developed a novel approach named ‘Emulsion PCR or EmPCR’ to overcome the problem of PCR duplicates. In Emulsion PCR every fragment is amplified in a single PCR reaction in an emulsion droplet. How?
Let’s find out.
In this article, I will explain to you the concept of emulsion PCR, its principle, how-to (process), advantages, applications and limitations. At Genetic Education Inc. we re-discover the knowledge using the simplest explanation. This article will add more value in your PCR learnings.
Stay tuned.
Key Topics:
What is Emulsion PCR?
To better understand the concept of emulsion PCR, we need to know the differences between PCR and emulsion PCR.
- PCR can amplify a single template DNA in a reaction, whereas emulsion PCR can amplify multiple (or many) template DNAs in a single reaction. Note that here template DNA is a target region on the DNA that we want to amplify.
- PCR amplifies the template in a single reaction, while in the emulsion PCR, many separate micro-amplification reactions for each template, occur in a single reaction.
- PCR causes duplication of a particular template, while emulsion PCR can amplify each template DNA, effectively.
- PCR can amplify a microliter (µL) DNA while the emulsion PCR can amplify picoliter (pL) DNA.
The emulsion PCR reaction occurs in water-in-oil emulsion droplets. Each individual droplet performs amplification for individual templates in a high-throughput reaction. It’s entirely different from the conventional PCR in which the reaction occurs in a solution.
Using physical means oil-to-water droplets are prepared, each droplet carries a single template fragment and reagents for amplification and performs the PCR reaction. Check out this image to better understand the concept.
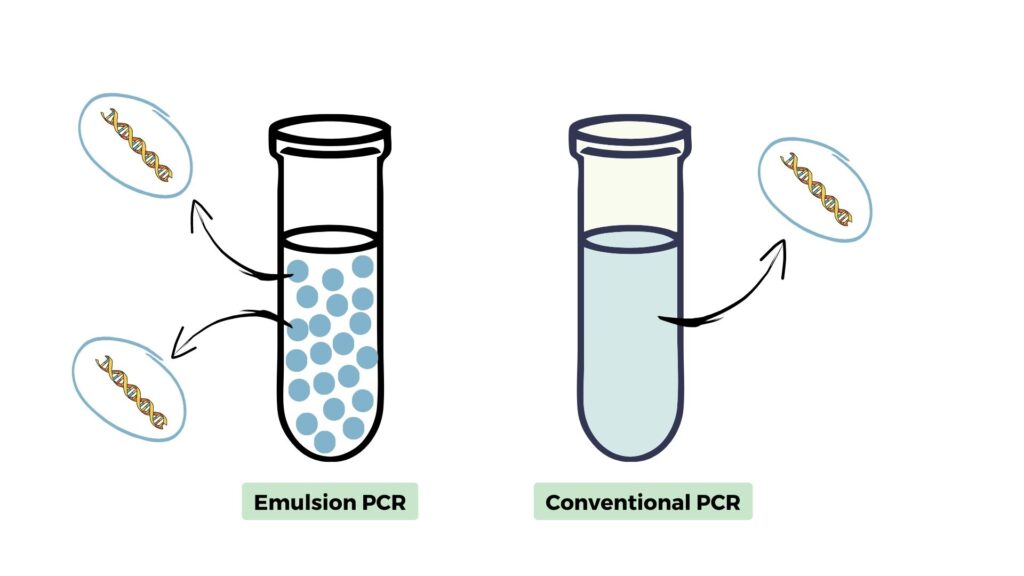
Principle of Emulsion PCR:
The principle of emulsion PCR states that the DNA template is partitioned into water-in-oil droplets and each separate DNA fragment is amplified independently in each droplet in a high-throughput reaction.
The entire reaction can be divided into 6 steps– DNA fragmentation, Emulsion formation, independent amplification, Breaking the emulsion, isolation of amplified DNA fragments and purification.
Steps of Emulsion PCR:
DNA fragmentation:
The template or genomic DNA is fragmented using the restriction enzyme into smaller-sized fragments and ligated to the known DNA adapters. Note that the adapters are short and known DNA sequences.
Emulsion preparation:
The emulsion for the emPCR is prepared by mixing the aqueous phase (buffer), oil phase— mineral or silicon oil and surfactant–Tween 20. The buffer or aqueous phase contains all the essential ingredients for the amplification.
When the aqueous phase, oil and surfactant are mixed, the emulsion droplets are created. Each droplet contains the ingredients (dNTPs, Taq DNA polymerase, primers and template DNA) for amplification.
PCR amplification:
Now each droplet containing ‘any’ individual fragment along with PCR ingredients (dNTPs, Taq DNA polymerase, primers and template DNA) is amplified when we run a standard PCR protocol.
Here, no need to design separate primer sets for each different fragment, as we already have added the known sequence adaptors.
Each individual droplet behaves like an independent reaction and amplifies the fragment present in the droplet. Through this, the chances of DNA duplication of a single type of fragment can be reduced.
Breaking the emulsion:
Now the next crucial step in this process is to break the emulsion droplet in order to release the amplified fragments. There are various physical and chemical methods available to break the emulsion, the choice depends on the requirement of the experiment.
Here, I have discussed a few popular emulsion-breaking methods.
Centrifugation:
Centrifugation is the best, easy and quick physical method for breaking the emulsion droplets. Upon completion of amplification, the sample is centrifuged at a higher speed. Amplified DNA fragments are released and separated into the aqueous phase.
High-speed centrifugation can disrupt the DNA structure.
Freezing and thawing:
Some literature suggests freezing/thawing for breaking the emulsion. This process can destabilize the water-in-oil droplets, break them and release the DNA.
Organic solvent extraction:
Yet another popular and common technique used in breaking the emulsion is the use of organic solvents like isopropanol or ethanol. An organic solvent destabilizes the chemical structure between the water and oil and thereby disrupts the emulsion. This eventually releases the DNA into an aqueous phase. The organic solvent-based technique has an excellent success rate and gives high-yield amplicons.
Use of detergents:
Some detergents may react with the surfactants, break the emulsion and release the amplified DNA.
PTT method:
Phase Transition Temperature is a technique in which the breaking of emulsion is subjected to temperature transition. For instance, temperature increment can destabilize the surfactant, water-in-oil structure and releases DNA fragments.
However, it’s important to understand that the higher temperature sometimes can denature the amplified DNA fragments as well.
Purification:
Once we collect the amplified DNA fragments into the aqueous phase, the amplicons should be purified before using them for sequencing or any other downstream applications. Techniques like alcohol precipitation, column-based purification or magnetic bead-based purification can be used to purify the amplicons.
Read this article to learn more about DNA purification: Methods for DNA purification.
This is a brief step-by-step procedure of emulsion PCR, the amplified fragments are stored in a DNA library, quantified and used for DNA sequencing.
How to use emulsion PCR in NGS:
Now in this section, I will explain to you the step-by-step process of how we can use the emulsion PCR in the NGS (Next-Generation Sequencing).
- Step 1: Isolate high-quality DNA using spin-column or automated DNA extraction. Measure the purity and quantity of DNA.
- Step 2: Fragment the DNA into smaller-sized fragments using restriction digestion.
- Step 3: ligate the known sequence adaptors and store them in the genomic DNA library.
- Step 4: add all the PCR ingredients into the buffer or aqueous phase.
- Step 5: Prepare the emulsion, using any of the techniques explained above.
- Step 6: Apply a standard PCR protocol and allow samples to be amplified.
- Step 7: Isolate the amplified DNA fragments using any of the techniques explained above.
- Step 8: Purify the DNA fragments as per the protocol.
- Step 9: Prepare a sequencing reaction and allow DNA sequencing.
- Step 10: Analyze the data generated.
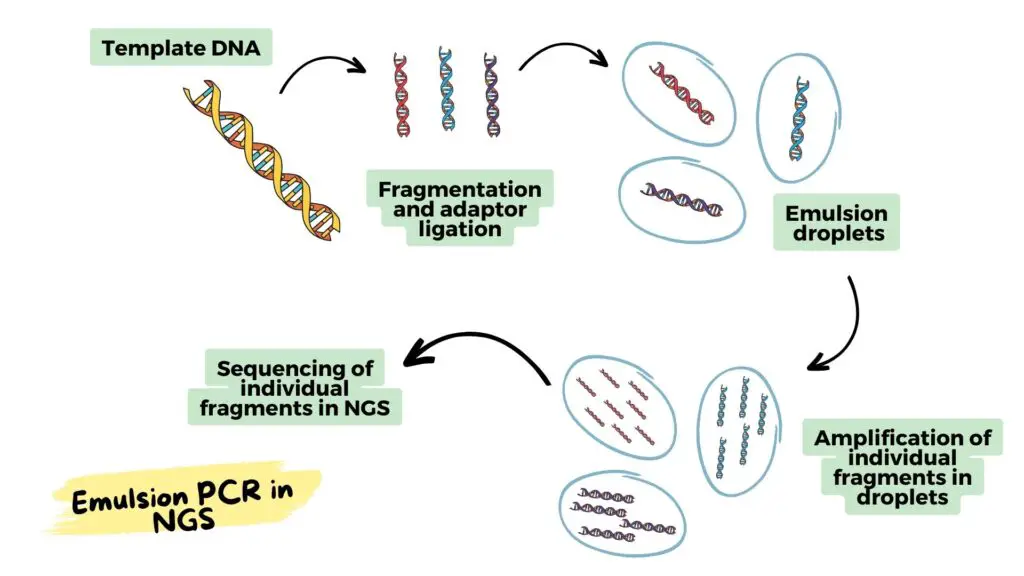
Advantages of Emulsion PCR:
Emulsion PCR has several crucial advantages over conventional PCR. Here are some.
- Firstly, it’s used to reduce PCR duplications by amplifying every single DNA fragment into individual reactions under the emulsion droplet.
- It provides a high throughput amplification of thousands to millions of DNA fragments in a single reaction and so is used as a tool for NGS library enrichment.
- As it can detect a few templates present in the sample, it’s highly sensitive, effective and accurate in amplification.
- It’s a cost-effective, simple and fast way to amplify DNA fragments for sequencing platforms like amplicon sequencing, NGS and targeted sequencing and genome sequencing.
- It’s a versatile, super easy, handy and highly precise technique.
Limitations:
Besides having excellent advantages in the genomic research field, scientists have to face some serious limitations.
- Emulsion preparation, maintenance and stabilization throughout the reaction is a complex, challenging and difficult process. In addition, it’s also a difficult and challenging task to prepare sufficient and uniform-sized emulsion droplets.
- The surfactants can interfere with PCR amplification and cause incomplete or partial amplification.
- It can produce emulsion PCR bias; it can produce varied-sized amplicons within the emulsion droplets.
- Another major problem scientists have to face during emulsion PCR is the merging of individual emulsion droplets. This can mislead the results.
- In addition, it also requires pre- and post-processing steps like emulsion preparation, amplicons separation and purification.
Applications of emulsion PCR:
Emulsion PCR is a specialized technique and not routinely used in a PCR lab, it’s indeed a subsidiary technique or we can say an optimization for NGS and single-cell genomic studies. Here are some of the important applications.
NGS:
Emulsion PCR is widely used for library enrichment in NGS (Next-Generation Sequencing). Thus, it provides a high-quality and high-yield NGS library for sequencing and thereby improves the accuracy and reliability of NGS.
Single-cell genomics:
Yet another fantastic application of emulsion PCR is in single-cell genomic studies to understand the differences and heterogeneity between individual cells. It can effectively amplify individual fragments of a single cell’s entire genomic DNA.
Detection of rare variants:
Emulsion PCR is often used in the selective detection of rare variants. A rare variant is technically a low abundant template DNA, meaning, present in a very low quantity. Thus, it can be missed during conventional PCR.
In this scenario, the rare template DNA is partitioned into individual emulsion droplets and amplified effectively.
Forensic analysis:
Emulsion PCR is an excellent tool for the detection of trace amounts of DNA obtained from any crime scene and therefore is nowadays widely used in forensic analysis.
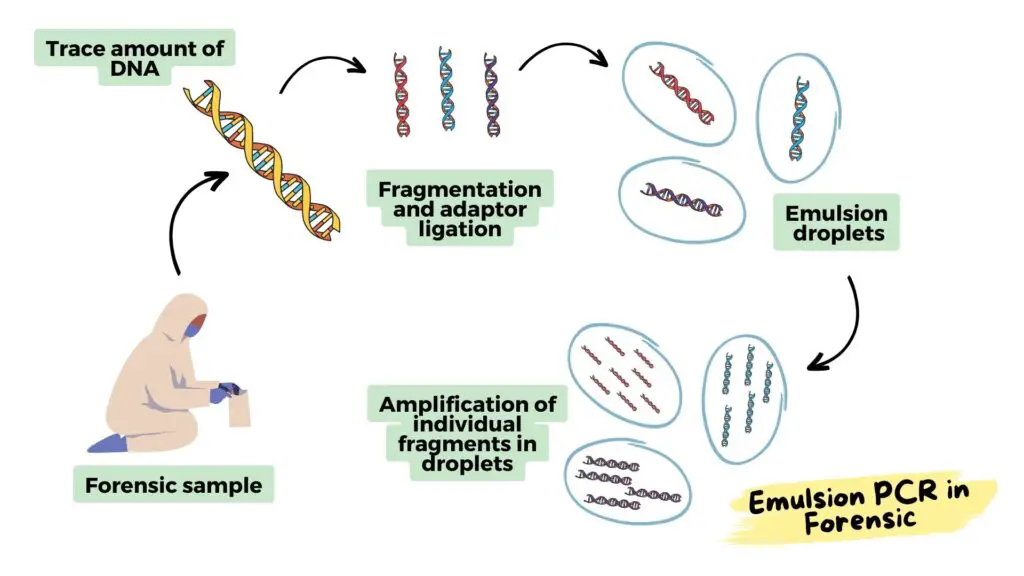
Besides these significant three applications, it can also be used in the droplet digital PCR for more accurate quantification, detection of mutations, copy number variations and environmental studies.
Wrapping up:
NGS and related techniques require a uniform and sufficient amount of DNA library and here, techniques like emulsion PCR plays a crucial role. Despite having some serious drawbacks, it is employed for NGS and other sequencing platforms due to this reason.
This is also important to generate uniformity in sequencing reads and coverage. I hope you understand the topic well. Comment below and let me know. If you are interested in PCR and related learning do not forget to participate in our online course.
FAQs:
Sources:
- Kanagal-Shamanna R. (2016). Emulsion PCR: Techniques and Applications. Methods in molecular biology (Clifton, N.J.), 1392, 33–42.
- Shao, K., Ding, W., Wang, F., Li, H., Ma, D., & Wang, H. (2011). Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PloS one, 6(9), e24910.