“High Throughput sequencing sequences many samples simultaneously in a single run. Learn about the concept of high throughput sequencing and why it is important.”
The initial sequencing of the human genome cost billions of dollars, but now the same process is available for just a few hundred dollars, thanks to advancements in sequencing throughput.
Sanger sequencing, which was the first-generation sequencing technique, was a pioneer during the Human Genome Project. It is certainly advantageous over other sequencing techniques available at that time, however, still has limitations.
Sanger comes with low throughput and high cost. The development of NGS- next-generation sequencing improved the throughput and reduced the cost of sequencing, greatly.
In this article let’s understand, what exactly is high throughput sequencing, how it works and why it is important. Let’s find out.
Related article: The Human Genome Project: Aims, Objectives, Techniques and Outcomes.
Key Topics:
What is High Throughput Sequencing?
Throughput simply means a process, method or technique that can handle multiple operations or samples in the same or less time. Technology advancements and robust analysis approaches help improve the throughput of any process.
High Throughput Sequencing (HTS) employs a more robust and advanced approach, allowing for the sequencing of various samples in a single run. By utilizing the same machine, chips, and chemicals for multiple samples simultaneously, HTS significantly reduces time, costs, and improves the Turnaround Time (TAT) for laboratories.
Previous generation sequencing techniques such as Sanger sequencing and Maxam-Gilbert sequencing were very low throughput methods. Contrarily, next-generation sequencing such as Pyrosequencing, Oxford nanopore sequencing and Illumina HiSeq, NextSeq are very high throughput, robust, efficient and accurate sequencing techniques available right now.
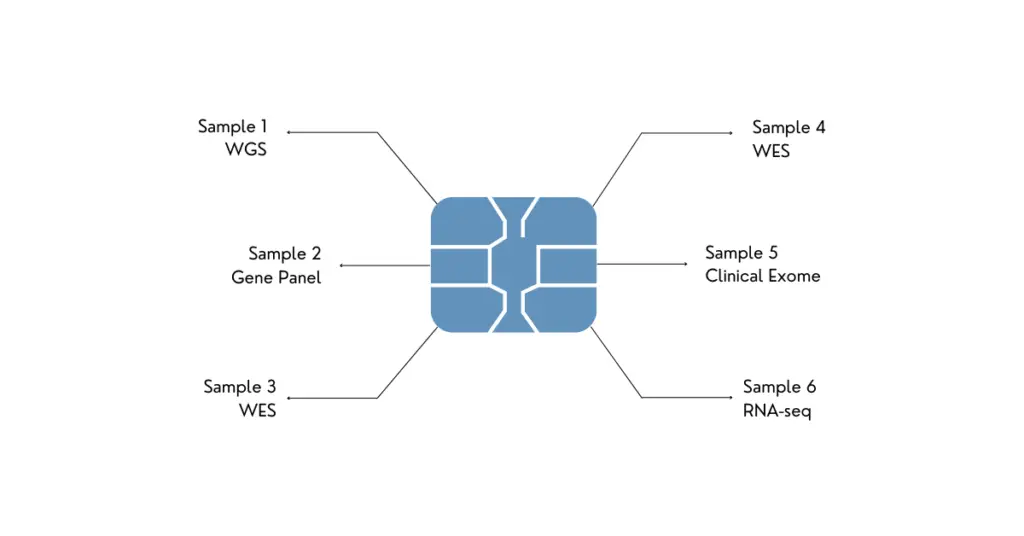
Why High Throughput Sequencing is Important?
Throughput is a critical parameter for any genome testing facility whether it’s research or clinical diagnosis. Let’s take a real lab example to understand it better.
Every day, a DNA sequencing lab has to test many types of samples, for example, whole-genome sequencing, transcriptome analysis, whole-exome sequencing, RNA sequencing and various gene panels for cancer.
For each sample and each ‘type’ of sample, setting and running separate sequencing procedures is a tedious, time-consuming and costly procedure. A high-throughput sequencing machine can run all these samples in a single run.
What a lab needs is an experienced molecular geneticist who can have such experience to run all these samples in a single run and handle the operations. The machine will run once for all these sample types and eventually save time and cost.
So high throughput reduces TAT (Turn Around Time) for a lab. However, a lab should have an excellent data processing facility to handle such a huge amount of data generated by high throughput sequencing.
Keep in mind that different software and pipelines are required to perform different types of analysis for different purposes. For example, Illumina’s NextSeq 500 series can process the whole-genome sequencing sample in 15 to 26 hours and generate 25 to 120 GB data per run for single and paired-end sequencing.
So HTS is important for research and diagnosis facilities because it generates a high amount of data for comprehensive analysis, reduces the TAT, runs time and cost per sample, and performs sequencing at high speed and efficiency.
High-Throughput Sequencing Techniques
Here is the comprehensive list of various next-generation high-through sequencing techniques available in the market.
| Sequencing Technique | Throughput | Run Time | Sequencing Strength | Chemistry | Number of Samples | Data Generated per Run |
| Roche 454 Pyrosequencing | Moderate to High | Hours to days | Long reads (400-800 bp) | Pyrosequencing | Single sample per run | Up to 1 Gb |
| HiSeq 3000 | High | Days | Short reads (100-150 bp) | Sequencing by synthesis (SBS) | Multiple samples per run (up to 384) | Up to 6 Tb |
| NextSeq 500 | Moderate to High | Hours | Short reads (75-300 bp) | Sequencing by synthesis (SBS) | Single to multiple samples per run | Up to 120 Gb |
| Oxford Nanopore | Variable | Real-time | Long reads (>100 kbp) | Nanopore sensing | Single sample per run | Varies (real-time sequencing) |
| ABI SOLiD sequencing | High | Days | Short reads (50-75 bp) | Sequencing by ligation (SBL) | Multiple samples per run | Up to 100 Gb |
| Illumina Solexa | High | Days | Short reads (50-300 bp) | Sequencing by synthesis (SBS) | Multiple samples per run | Up to 6 Tb |
How Does HTS work?
In this section, we will understand various steps for high throughput sequencing. High throughput and next-generation sequencing platforms, although complex, comprise similar steps, excluding the chemistry.
Step 1 is nucleic acid isolation. Here a high-quality and quantity of nucleic acid is isolated from the given sample.
Step 2 is Library preparation. The whole genome can not be sequenced in a single run. So to achieve efficient sequencing, DNA fragments are generated to prepare fragment libraries.
Step 3 is adaptor ligation. Now, known sequence adaptors have been ligated with each fragment. Adaptor ligation serves a known point location during assembling. Note that index and sequencing primer sequences are also added.
Step 4 is library enrichment. On some platforms, the libraries are amplified in an amplification reaction before sending the sample for sequencing. It generates an adequate amount of fragments for efficient sequencing.
Step 5 is sequencing. Now, a sample chip or flow cell is loaded into the machine. From machine to machine and company to company, sequencing chemistry varies. For example, in Illumina sequencers, the machine reads sequences using sequencing by synthesis chemistry.
The chemistry of various instruments is listed in the above table. We will also discuss each chemistry in separate articles.
Steps 6 and 7 are data analysis and interpretation. Various bioinformatics pipelines and software are used to analyze the high-throughput sequencing data analysis. An expert analyzes the data, interprets it and prepares results.
Related articles:
- A Beginner’s Guide to Sanger Sequencing Results [Before Electropherogram Analysis]
- 4Peaks Review: Easiest Sequence Analysis Software
- Advantages and Limitations of Sanger Sequencing
- What is NGS?- Definition, Principle, Steps, Chemistries, Advantages and Limitations
- De Novo Sequencing: Steps, Procedure, Advantages, Limitations and Applications
Wrapping up
In conclusion, high throughput sequencing platforms, which are now known as next-generation sequencing have revolutionized the field of genetic research, disease diagnosis and personalized genomics.
Now, scientists can sequence the whole genome within a day which took years during the human genome project. Still, the overprice of the sequencing setup is a major hurdle for clinicians, labs and research organizations to acquire NGS in their labs.
I hope your fundamentals for high throughput sequencing are now clear. Do share this article and bookmark the page.
Sources:
Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015 May 21;58(4):586-97. doi: 10.1016/j.molcel.2015.05.004. PMID: 26000844; PMCID: PMC4494749.


