Replication is a process to manufacture a new DNA, a duplex DNA makes another duplex. DNA polymerase actively participates in this process and controls the whole from start to end.
Replication often said as “DNA synthesis” or “DNA copying” is an essential process for life. Making and transmitting similar DNA is the key function, replication has. And in a normal time, it does do it correctly and accurately.
It’s a catalytic multienzyme process including many different enzymes in each step because so much work is to be completed during a normal replication cycle.
dsDNA opening, ssDNA strand holding, primer binding, releasing tension and adding nucleotides, all enzyme govern processes that occur simultaneously and rhythmic.
However, the major part of the process “replication”- to add correct nucleotides is performed by DNA polymerases. Meaning, couple of different DNA polymerases catalyze various steps and reactions.
What it normally does do is incorporate complementary bases to the template DNA, using the free 3’-OH end of the RNA primer. By forming phosphodiester bonds between nucleotides, it makes another strand.
The whole process occurs in a fraction of a second and therefore error may occur during. In a normal cell, 1 mismatch at every 1000000000 nucleotide addition is reported.
But the cell doesn’t even tolerate that much error and tries to repair it. A whole set of proteins, enzymes and factors actively engage in the process of correcting the replication error- known as DNA “proofreading.”
The fidelity of DNA replication highly relies on the activity of DNA polymerase.
Key Topics:
What is DNA proofreading and how does it occur?
In biology, the “proofreading” process was first explained by John Hopfield and Jacques Ninio.
Proofreading is a process of checking if replication or base-pairing occurs complementary or not. Although the error rate in replication is 1 into 1000000000. Correct base pairing A with T and G with C should occur precisely.
Wrong base-pairing replaces the original nucleotide with the wrong one and transmits it to offspring too. Meaning, it produces mutation.
Proofreading by DNA polymerase and exonucleolytic proofreading are two common mechanisms that work finely. Proofreading and correct base pairing occur either during replication or after the completion of replication.
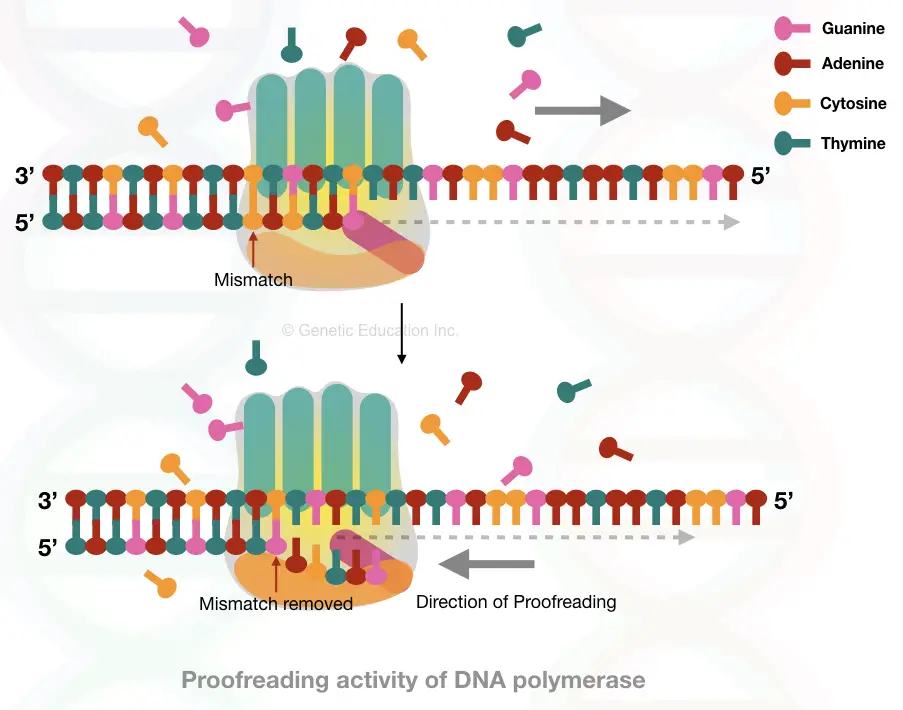
DNA polymerase in proofreading:
During the replication, DNA Pol III does do the task (in prokaryotes). It has two kinds of activities viz. Polymerase and exonuclease. While incorporating nucleotides, the polymerase closely monitors correct base pairing and it generally performs it finely.
If any wrong nucleotide is incorporated, it immediately identifies it, gets back to it, breaks the phosphodiester bond, removes the mismatch and incorporates another correct nucleotide.
DNA polymerase adds nucleotides as well as removes incorrect nucleotides too. When nucleotide triphosphates are coming, polymerase chooses only the correct one and adds it to the template DNA.
The wrong base has a low binding affinity, polymerase undergoes conformational changes and dissociates it. Once it assures the correct binding, it moves ahead.
Sometimes polymerase even can’t recognize and correct the wrong base pairing, however, exonuclease (which is a domain of polymerase) identifies these weak bindings and helps to correct them.
Here is the list of prokaryotic and eukaryotic polymerases having polymerase and exonuclease activities.
| DNA polymerase | Polymerase activity
(5’ to 3’) | Exonuclease activity
(3’ to 5’) | Exonuclease activity
(5’ to 3’) |
| Prokaryotic DNA polymerases | |||
| I | Yes | Yes | yes |
| II | Yes | Yes | No |
| III | Yes | Yes | No |
| IV | Yes | Yes | No |
| V | Yes | No | No |
| Eukaryotic DNA polymerases | |||
| Alpha | Yes | No | No |
| Beta | Yes | No | No |
| Gamma | Yes | Yes | No |
| delta | Yes | Yes | No |
| epsilon | Yes | Yes | Yes |
The exonuclease activity of the polymerase helps to break the phosphodiester bond between nucleotides and removes the wrong one. Immediately, the polymerase activity of the DNA polymerase adds the right nucleotide. Unlike other polymerases, only DNA polymerase III has this special kind of power to perform both functions in prokaryotes during replication.
If mismatch still remains in the newly formed DNA, another proofreading will occur shortly after the completion of Replication. Specialized protein identifies the nick at wrong base pairing; again, the exonuclease activity of polymerase removes the mismatch.
MutS and MutL are proteins that work as a “mechanic” at the site of error or wrong base pairing. When an error occurs the MutS finds it, slips back towards the site and stays there strongly. Immediately, the MutL finds the site (where the MutS locked) and creates a nick there. Polymerase does do its part, soon after.
The polymerase part incorporates the right nucleotide. The ligase seals the gaps by creating a phosphodiester bond. Although, how proteins find the mismatch between parental and newly formed DNA is still not well understood, especially in eukaryotes.
In prokaryotes, the DNA methylation process helps to recognize the wrong pairing. For example, in E Coli, shortly after completing replication, methylation occurs at the adenine base of the parental strand but not on the newly formed strand.
Lack of methylation on the daughter strand is identifiable by the polymerase using which; it finds the mismatch and repairs it.
We explained that methylation in prokaryotes helps distinguish parental strand from daughter strand. But that’s not the case in eukaryotes. Then how does discrimination occur?
The strand discrimination signals are provided by the PCNA protein. The PCNA remains there during replication and gives signals, which DNA strand is new.
Subsequently, the MutS, MutL and DNA polymerase works accordingly. Notedly, three hypotheses are proposed to explain this.
- First, the MutS and MutL form a clamp and slide toward the signals given by the PCNA.
- Second, make a fold in DNA and attract PCNA towards.
- Third, mark the sliding area to delete it.
Studies show that among all three hypotheses, the third one is somehow correct. Once the MutS is placed on the site, many MutL proteins gather at the site and bind to the whole place to be repaired.
During the whole process, both MutS and MutL do conformational changes in their shape to give further signals and recruit other proteins and enzymes and prevent packaging in this region.
Weninger and Eric further explained that during the process of repair, both proteins remain there, and protect the strand from conformational and undesirable changes. Meaning, prevent further DNA packaging.
Eric is a professor and member of UNC-chapel hill, Lineberger. Weninger K is a biophysicist and university faculty scholar at NC state university. Both are the authors of the present work.
Several DNA mismatch repairs also occur when damaged by extrinsic factors such as UV light. It usually adds wrong base-pairing and forms dimers between the same adjacent nucleotides.
A team of diagnostic experts (the exonuclease and other proteins) finds the dimer, cleave on both the ends; 3’- OH and 5’-P, remove the whole part which is not normal and have the mismatch.
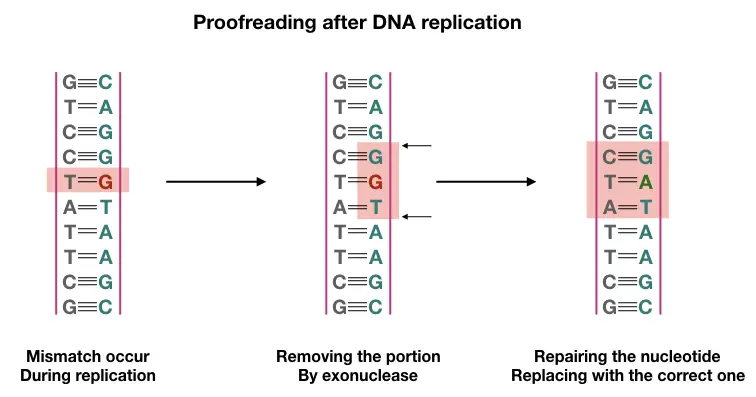
The polymerase then binds to the site and inserts correct nucleotides again. The ligase seals gaps by forming a phosphodiester bond. The mechanism is known as ‘nucleotide excision repair.’
In prokaryotes, especially in bacteria, all three DNA polymerases; viz DNA pol I, DNA pol II and DNA pol III have both polymerization and exonuclease activities. Contrary, in eukaryotes, only DNA polymerase epsilon have that activity.
Usually, polymerase and exonuclease activity occurs in the 5’ to 3’ and 3’ to 5’ direction, respectively.
Read more: Prokaryotic DNA Replication: Initiation, Elongation and Termination.
4 ways to DNA proofreading:
- During polymerization, the polymerase moves back, removes and inserts the wrong nucleotide and correct nucleotide pairing, respectively.
- Through the exonuclease activity of DNA polymerase. The exonuclease domain cleaves off the wrong nucleotide and adds a new one.
- Though mismatch repair. After the completion of replication, the organized activity of polymerase and exonuclease do proofreading.
- Through nucleotide excision repair. After the completion of replication, Enzyme polymerase with the exonuclease activity and ligase do the proofreading.
The complete process of all four pathways is explained in the above sections. And the function of various prokaryotic and eukaryotic polymerase has explained here:
| DNA polymerase | Function |
| Prokaryotic DNA polymerases | |
| I | Remove and replace prime |
| II | DNA repair |
| III | DNA replication and elongating strand |
| IV | DNA repair |
| V | DNA repair and translesion DNA synthesis |
| Eukaryotic DNA polymerases | |
| Alpha | Primer synthesis and repair |
| Beta | DNA repair |
| Gamma | Mitochondrial DNA replication |
| delta | Lagging strand synthesis and repair |
| epsilon | Leading strand synthesis and gap filling. |
What if not proofreading?
The objective of performing proofreading is to transmit the correct DNA sequence to daughter cells, regulate genetic integrity, and maintain genetic continuity. In addition, lowering the mutation rate is also an important objective of DNA proofreading.
Lack of proofreading produces mutation and transmits it to other cells. Deletion and insertion are common errors in replication. Inheriting mutation is a bit tricky process, I can say.
Lack of proofreading replaces the whole nucleotide or adds a brand new nucleotide to the newly formed strand. Let’s take an example to understand.
Suppose if a wrong base “G” is added while replicating the daughter DNA. The correct nucleotide on the parental strand is the A. So technically it should be “T” instead of G.
If the “G” isn’t corrected, during the second round of replication, the newly formed strand has a C nucleotide instead of T. See the image below, you can better understand.
Cancer is a common product of an error in proofreading. Lack of exonuclease activity is reported in colorectal cancer. Lynch syndrome is a kind of cancer that occurs due to many replication errors during cell division.
Wrapping up:
Replication is an important process to sustain and survive. New DNA forms only when it occurs. New mutations too originate from it. Mutations commonly aren’t helpful.
If the mutation is useful it can be transmitted to offspring and spread to the population. Once again, replication spread it. However, to maintain the integrity of life and avoid unwanted changes, DNA proofreading continuously works.
Proofreading reduces the rate of mutations a thousandfold.
Source:
North Carolina State University. “How DNA ‘proofreader’ proteins pick and edit their reading material.” ScienceDaily. ScienceDaily, 21 August 2015.