“NGS platforms highly rely on fluorescently labeled nucleotides. However, different fluorophore positions influence NGS speed, precision and efficiency.”
NGS chemistries vary among different platforms, with fluorescence and proton/semiconductor detection being the most common methods. Illumina and PacBio use fluorescence detection, while Ion Torrent and ION use semiconductor detection.
However, fluorescence detection is the most popular, efficient and accurate chemistry for large-scale and high-throughput sequencing. It typically involves four different fluorescent tags, or sometimes just two and their combinations, to identify nucleotides during sequencing.
The software captures the fluorescence released from the sequencing by synthesis and decides which nucleotide is incorporated in the sequence depending on the color emission.
Interestingly, where the fluorescent tag is attached to a nucleotide has a significant impact on the NGS speed, precision and accuracy. In this article, I will majorly discuss why and how differential nucleotide labeling influences NGS performance.
Stay tuned.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Key Topics:
What is nucleotide labeling in NGS?
NGS platforms usually follow the sequencing by synthesis scheme for efficient sequencing. During the process, the polymerase picks a complementary nucleotide and incorporates it into the single-stranded DNA template.
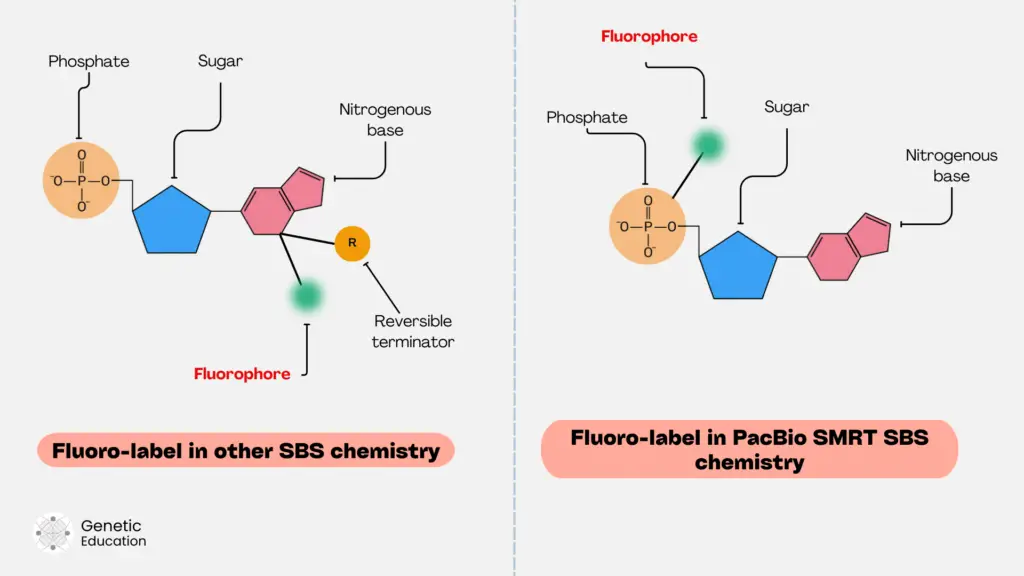
Labeling nucleotides with different fluorophores helps detect the incorporation. To do so, a fluorophore is attached to any location on a nucleotide. Either on a sugar, nitrogenous base or phosphate.
Nucleotide labeling types in NGS
Keep in mind that a nucleotide being incorporated in a sequence is made up of sugar, phosphate and a nitrogenous base.
Illumina platform uses fluorescent nucleotide labeling on the nitrogenous base. Along with that, the reversible terminator is incorporated into the sugar molecule of the nucleotide.
In another chemistry, both the reversible terminator and the fluorophore are attached in the same position on a nitrogenous base.
A fluorophore can also be attached to the pentose sugar of a nucleotide, but unfortunately, no NGS chemistry uses this type of labeling.
PacBio uses innovative nucleotide labeling in which a fluorophore is attached to the phosphate. These are the three common types of fluorescent nucleotide labeling procedures followed by various NGS platforms.
How is nucleotide labeling used in NGS?
Now the important question is, how does the detection occur? Or sequence read?
Whenever a nucleotide analog is incorporated at its complementary location during the sequencing procedure, the fluorophore is removed from the nucleotide. Because polymerase has to add the exact complementary nucleotide in the sequence.
When a fluorophore is attached to the nucleotide, the polymerase considers it as a nucleotide analog and not the correct nucleotide. So it removes the fluorophore, upon nucleotide addition.
Now, the fluorophore releases fluorescence. A high-quality camera captures the light snap while the signals are detected by the detector. The software converts the fluorescent signal into a digital sequence format based on the type of fluorescence released.
Impact of nucleotide labeling on NGS performance
Now, the process is looking simple and effective, isn’t it? But what’s the catch?
The sequencing by synthesis process, which is the heart of almost every NGS platform, mimics the natural replication process. Whenever it finds a complementary “nucleotide” it adds to the growing DNA strand.
But in sequencing the fluorophore-labeled nucleotides are not true nucleotides, those are the nucleotide analog or we can base analog. Now, here is the problem!
Polymerase is an enzyme that follows a simple lock and key model. It can only work if it finds the exact ‘key’, meaning, it can incorporate a complementary nucleotide only if it finds a correct nucleotide.
Polymerase took time to recognize the nucleotide analog, identify it, remove the extra fluorescent tag and then incorporate the nucleotide. In addition, the use of a reversible terminal in the case of Illumina sequencing makes the process even slower.
So overall, the fluorophore labeling at nitrogenous base or sugar, makes the overall polymerization process slower. Thus, the short read sequencing (Illumina) is a bit slower process.
Conversely, in the case of PacBio sequencing the fluorophore labeling is known as phospho-linked nucleotide labeling, the fluorophore is attached to the phosphate. That acts as a natural nucleotide.
Because the phosphate is ultimately released during the nucleotide incorporation process in replication and in in vitro SBS, both the process. So along with that, the fluorophore is naturally removed, releases fluorescence and is detected.
PacBio uses Hifi sequencing technology. That means, it uses Hi-fidelity DNA polymerase during the synthesis process. The HiFi polymerase can add up to 10 nucleotides in a second.
So using ‘natural-kinda’ labeling nucleotides, instead of base labeling nucleotides, speeds up the overall process. This will eventually increase the precision and efficiency of sequencing.
Hence, phospholinked fluoro-labeled nucleotides used in PacBio long-read sequencing increase the sequencing speed for a read. And that’s why it’s known as Hi-Fi reads.
Wrapping up
Using fluorophore-labeled nucleotides in NGS platforms has pivotal importance, as it increases the detection accuracy and speed. Unfortunately, the sequencing accuracy for long-read sequencing is comparatively less, even though it uses the most efficient nucleotide incorporation mechanism.
There are many reasons for that, but the most common one is the number of reads generated. Short read sequencing generates way more reads than long read sequencing and that will give us more confidence in sequencing assembling.
Thus PacBio’s long-read sequencing is faster but comparatively inaccurate than other sequencing platforms. One study suggests that the SNP to wrong SNP detection rate, error rate, and wrong base calling rate are higher (<Q10) in the case of PacBio sequencing.
I will provide the article here, you can read and verify the information. Share this article and subscribe to our blog.
Sources:
Quail, M.A., Smith, M., Coupland, P. et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341 (2012). https://doi.org/10.1186/1471-2164-13-341.