“Sanger sequencing is a genetic technique to determine the order of nucleotides while PCR generates copies of the DNA fragment, present in the sample. Let’s see some of the significant differences between both methods.”
PCR and Sanger sequencing are two important and widely used techniques in genetics. Scientists can perform DNA fingerprinting, genetic tests, screen genetic diseases and study mutations. In addition, both have a significantly important role in gene therapy and genetic engineering too.
Both techniques are different and have their own importance, mechanism of working, principle, advantages and limitations. In this article, we will compare the two most important genetic techniques- Sanger sequencing and PCR.
Stay tuned.
Key Topics:
Sanger sequencing vs PCR
In this section, we will discuss some of the common as well as technical differences between both techniques.
Sanger sequencing is a DNA sequencing technique while PCR is an amplification technique. Sequencing reads each nucleotide while PCR amplifies each nucleotide present in the sample.
In 1977, Frederick Sanger postulated the first and foremost sequencing technique while in the year 1983, Kary Mullis postulated the PCR technique.
The working principle of Sanger sequencing is “chain termination.” When the polymerase adds any of the ddNTPs instead of a dNTP, the chain terminates and gives a signal. Conversely, the working principle of PCR is a polymerase chain reaction in which polymerase adds dNTPs to the growing DNA strand.
Hence only dNTPs are used in the PCR while dNTPs and ddNTPs both are used in the Sanger sequencing.
Chain termination occurs on a single strand of DNA and therefore only a single primer is used during Sanger sequencing, contrary, a pair of primers– forward and reverse primers amplifies two different DNA strands during PCR.
The ultimate goal of Sanger sequencing is to read the sequence and therefore only a single primer is required, on the other hand, the ultimate goal of PCR is to generate copies of DNA and therefore a pair of primers are used to amplify two different DNA strands and generate more copies.
Sanger sequencing results analysis requires poly acryl amide gel electrophoresis or capillary gel electrophoresis and additional computational software and tools. However, conventional PCR results analysis requires only a native agarose gel electrophoresis.
No additional computational tools PCR needs. In addition, qPCR even can’t need gel analysis as it runs and analyze results in a machine.
Sanger sequencing can read every nucleotide present in a sample and give us an idea about the structure and order of the nucleotide present in the sample. For this entirely different system, a fluorescent detector and signal recorder camera are used.
On the other side, PCR is a simple machine that can’t read nucleotides but amplifies them. It lacks parts like a detector or recorder. A single heating coil generates temperature differences for each step in a cyclic manner.
So the power of Sanger sequencing allows us to find out new sequence variations if present in the sample, nonetheless, PCR can’t find any new sequence variation or can’t even read it. PCR just validates the pre-existing sequence variations.
No prior sequence information is required for Sanger sequencing. But to run any PCR prior-sequence information is needed to design primers, set annealing temperature and target the mutation.
The chemistry behind PCR amplification is a temperature-based (thermal amplification) amplification while the chemistry behind sequencing is fluorescence detection. Sequencing needs additional fluorescence labeling and detection system.
Sanger sequencing can effectively address single nucleotide polymorphism and therefore is more sensitive and useful. On the other side, PCR can’t address or find single base change effectively, however, only a few modified updations are available for only a few mutations to validate the change.
The technical requirements for individual techniques are also different.
Sanger sequencing needs library preparation. Longer gene segments are first fragments into readable length, enriched and ligated with an adapter to prepare the library. In the very last step, the library is purified before sequencing. PCR doesn’t need such extension pre-preparation, besides only PCR reaction preparation.
Yet another technical difference between both techniques must be discussed regarding the primers used.
In both the techniques, the primers used are DNA primers, however, in PCR we need sequence-specific primers– it should have a binding site on the target sequence, while in the Sanger sequencing, it isn’t necessary. Here the primer is specific to the adapter, where they bind and initiate sequencing reactions.
Steps in each technique are enlisted here.
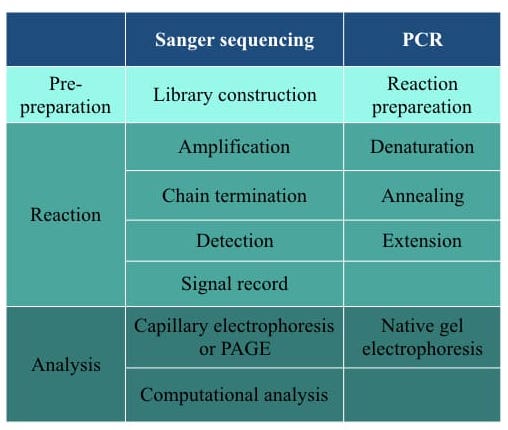
Nowadays, automated and semi-automated Sanger sequencing platforms are available which makes them more robust. However, PCR is still a semi-automated method in which the entire interpretation part has been carried out manually which makes it less robust.
Less human input is required to interpret the results of Sanger while the entire PCR part needs expertise and experience (more human input) to evaluate the results.
Advantages and limitations
Sanger sequencing is a fully or partially automated, robust, sensitive and reliable technique. It can read short sequences effectively, however, it needs extensive sample preparation and precision. The technique is costlier.
PCR is simple and we can say partially automated technique. It can detect a few known mutations and is a cost-effective option. On the downside, PCR isn’t fully automated and need additional requirement to read and interpret results.
Applications of sequencing
- SNP and indel genotyping.
- Find common, known and unknown sequence variations.
- Study a gene or its part.
- 16 and 18S rRNA gene sequencing.
- DNA fingerprinting.
- HLA typing.
- Finds mutations and genes associated with a disease.
Applications of PCR
- Study single gene disorders.
- Study known genetic variations.
- DNA fingerprinting.
- Validation of insert.
- Site-directed mutagenesis.
- Library preparation.
- Sample enrichment.
Read this article: 50 powerful applications of PCR.
Quick summary:
| Sanger sequencing | PCR |
| Read every nucleotide present in the sequence | Amplifies every nucleotide present in the sequence. |
| The technique was developed by Sanger F in 1977. | The technique was developed by Kary Mullis in 1983. |
| Generate a readable form of sequence. | Generates multiple copies of a sequence. |
| The working principle is, “chain termination.” | The working principle is, “thermal amplification.” |
| Use dNTPs and ddNTPs (ddNTPs terminates the synthesis). | Use only dNTPs during the synthesis. |
| Requires only a single primer. | Requires a pair of primers. |
| The primer should not be specific to the target sequence. | Primers must be specific to the target sequence. |
| Pre-preparations like DNA fragmentation, adapter ligation and library preparation are required. | Only reaction preparation is required. Lacks extensive pre-preparations. |
| Sequence information doesn’t require | Need sequence information to prepare the assay. |
| Use fluorescent chemistry. | Don’t use fluorescent chemistry. |
| Requires additional setup like fluorescence detector and recorder. | Only a simple thermocycler is required. |
| Entire technique is robust and automated. Sequencing, reading and evaluation are automated. | Needs additional techniques like electrophoresis to read and study results. |
| Additional computational software/tools are needed. | No additional computational tools are required. |
| Can read new sequence variations. | Can’t read new sequence variations. |
| Can provide novel sequence information | Can’t provide novel sequence information. |
Wrapping up:
In conclusion, Sanger sequencing has won the beetle against PCR as it’s more robust, sensitive and detection power. The main advantage of Snager is its ability to find new sequence variations. While PCR can be used at the research level and study inherited diseases (single gene).
Known sequence variations can be addressed by PCR. In recent times, researchers are more interested to study sequence variations rather than just validating them. Nonetheless, PCR is an un-replaceable technique in genetics and is required during almost every experiment. Even during the sequencing too.
I hope you like this comparative analysis article. Please share this article and bookmark the page.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.