“Structural chromosomal abnormalities occur due to deletion, duplication, translocation or inversion. Learn the concept, mechanisms and genetic abnormalities associated with structural chromosomal aberrations.”
Chromosomes carry genetic information in the form of DNA, which is known as the fundamental unit of heredity. There are 23 pairs of chromosomes in humans, including 22 pairs of autosomes and 01 pairs of sex chromosomes.
Chromosome number and structure play critical roles in the development and normal functioning of genes in humans. However, the chromosome number remains unchanged for an organism and varies among different organisms.
Any change in the chromosome structure or numbers causes serious abnormalities. Such changes are categorized into genetic mutations.
There are two types of chromosomal anomalies: numerical and structural. Numerical chromosomal aberrations or aneuploidies arise due to the changes in the number of chromosomes present in the cell.
On the other hand, when the structure of the chromosomes changes, it gives rise to structural chromosomal aberrations, which lead to several genetic disorders.
Structural aberrations include translocations, deletions, duplications, inversions, insertions, and many more. In this article, I will explain structural chromosomal anomalies, their mechanisms, and associated genetic conditions.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy to understand manner for better comprehension. A complete list of sources is provided after the article for reference.
Key Topics:
Definition:
Changes in the physical structure of the chromosomes such as deletion, duplication, translocation or inversion are known as structural chromosomal abnormalities.
This type of aberration involves changes in the architecture of the chromosomes, meanwhile, aneuploidies such as trisomy and monosomy involve variations in the number of chromosomes.
These differences are the results of errors that occur during DNA replication, DNA repair, and recombinations.
Certain physical factors also contribute to these types of aberrations, such as exposure to mutagenic agents, including radiation and chemicals. These aberrations can affect any chromosome, which leads to genetic disorders. The severity of the disease depends on the genes involved and the extent of the structural variation.
Types:
According to the nature of the structural change, structural aberrations can be classified into several types. Some of the main types are listed below:
- Translocations
- Deletions
- Duplications
- Inversions
- Ring Chromosomes
- Isochromosomes
Each type of abnormality has distinct characteristics and consequences, which are discussed in brief below.
Translocations:
A translocation involves the rearrangement of parts of the chromosome, which can be defined as “A Piece of chromosome breaks and reattaches with a different chromosome” or
“Exchange of parts of chromosomes between non-homologous chromosomes”.
Common types of translocations are balanced (don’t alter the amount of genetic material), unbalanced (Alter the amount of genetic material), reciprocal (exchange of segments between two chromosomes), and Robertsonian translocations (fusion of two acrocentric chromosomes at the centromere).
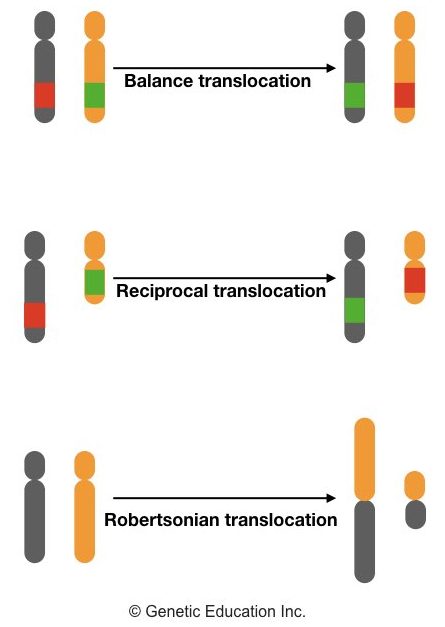
Mechanisms of Translocation:
Translocations occur due to errors during the recombination process. Reciprocal translocations arise when breaks occur in two different chromosomes, and the parts of chromosomes are exchanged.
Meanwhile, in Robertsonian translocations, two acrocentric chromosomes fuse at the centromere, resulting in a single chromosome.
Consequences of Translocations:
The consequences of translocations depend on the genes being translocated. Translocations of important genes or regulatory regions can lead to several genetic disorders. While translocations that do not affect the genes remain asymptomatic.
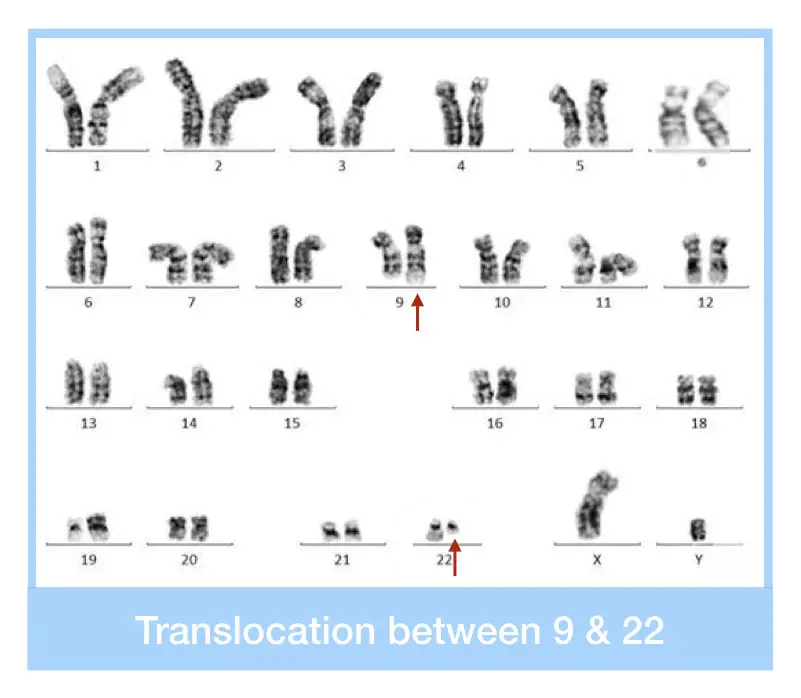
Examples of structural aberrations due to translocations, including chromosomes involved, symptoms, and severity, are listed in the table below:
| Name of Condition | Chromosomes Involved | Symptoms | Severity |
| Chronic Myeloid Leukemia (CML) | t(9;22) (Philadelphia chromosome) | Fatigue, weight loss, enlarged spleen, excessive white blood cell production | Severe (cancerous condition) |
| Down Syndrome (Translocation Type) | t(14;21) or t(21;21) | Intellectual disability, distinctive facial features, heart defects | Moderate to severe |
| Acute Promyelocytic Leukemia (APL) | t(15;17) | Bleeding, fatigue, susceptibility to infections | Severe (cancerous condition) |
| Ewing Sarcoma | t(11;22) | Bone pain, swelling, fractures, fatigue | Severe (cancerous condition) |
| Burkitt Lymphoma | t(8;14) | Swollen lymph nodes, abdominal pain, fever, night sweats | Severe (cancerous condition) |
| Synovial Sarcoma | t(X;18) | Swelling, pain, limited mobility in joints | Severe (cancerous condition) |
| Rhabdomyosarcoma | t(2;13) or t(1;13) | Muscle swelling, pain, lumps, vision problems | Severe (cancerous condition) |
| Follicular Lymphoma | t(14;18) | Swollen lymph nodes, fatigue, weight loss | Severe (cancerous condition) |
| Papillary Thyroid Carcinoma | RET/PTC rearrangements (e.g., t(10;17)) | Thyroid nodules, hoarseness, difficulty swallowing | Moderate to severe |
| Acute Lymphoblastic Leukemia (ALL) | t(12;21) or t(9;22) | Fatigue, frequent infections, bruising, bone pain | Severe (cancerous condition) |
Deletions:
Chromosome deletion is when a part of the chromosome is deleted. This results in structural chromosome rearrangements, bringing two separate genes together. This can result in the loss of one or more genes, leading to a variety of genetic disorders.
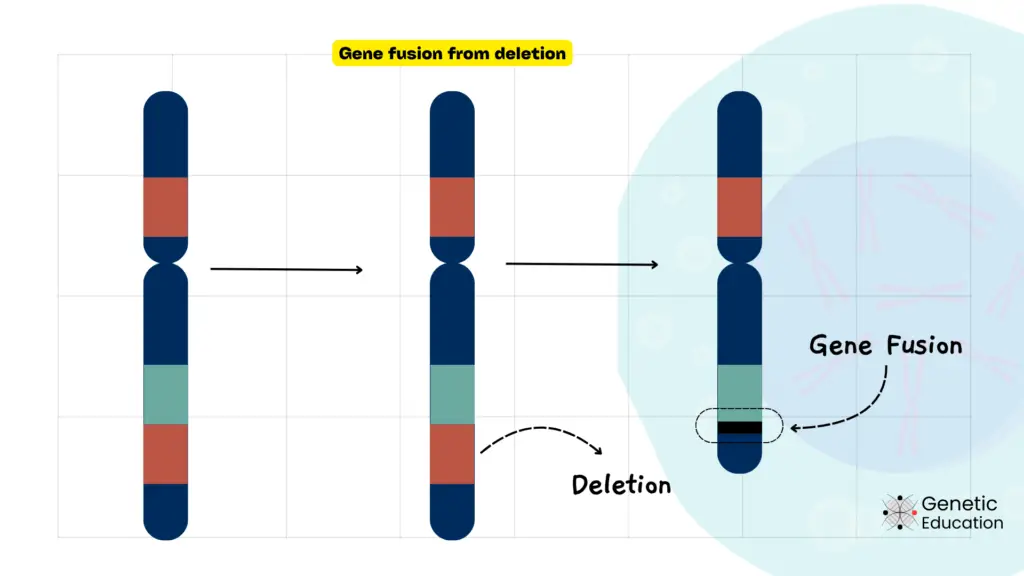
Mechanisms of Deletion:
Chromosomal deletions occur due to unequal crossing-over during meiosis, leading to the subsequent loss of a chromosomal segment, errors during DNA replication, and DNA repair.
It also occurs due to mutagenic exposure such as radiation, chemicals, or carcinogens.
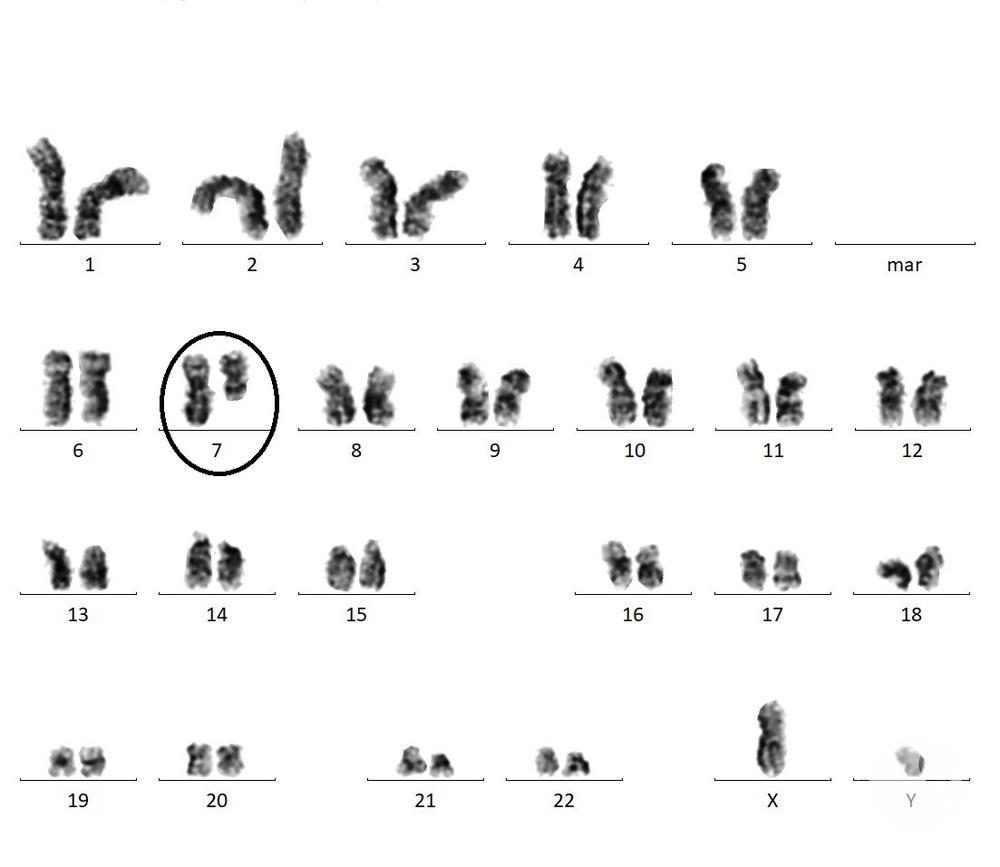
Consequences of Deletions:
The size and location of deletions in the chromosomes are the key factors when discussing the consequences of the deletions. When larger segments of the chromosomes are deleted, they are more likely to result in severe genetic abnormalities because this can involve multiple genes.
On the other hand, deletions of small segments lead to abnormalities that can be mild to severe. Deletions in the non-coding regions are mostly asymptomatic.
Examples of structural aberrations due to deletions, including chromosomes involved, genes involved, symptoms, and severity, are listed in the table below:
| Name of Condition | Chromosome Involved | Genes Involved | Symptoms | Severity |
| Cri-du-chat Syndrome | 5p (short arm of chromosome 5) | CTNND2, TERT, SEMA5A | High-pitched cry, intellectual disability, distinctive facial features | Moderate to severe |
| Williams Syndrome | 7q11.23 (long arm of chromosome 7) | ELN, LIMK1, GTF2I | Cardiovascular disease, developmental delays, “elfin” facial features, friendly personality | Mild to moderate |
| Prader-Willi Syndrome | 15q11-q13 (long arm of chromosome 15) | SNRPN, NDN, MAGEL2 | Obesity, intellectual disability, behavioral problems, hypotonia (low muscle tone) | Moderate to severe |
| Angelman Syndrome | 15q11-q13 (long arm of chromosome 15) | UBE3A | Developmental delays, seizures, lack of speech, happy demeanor | Moderate to severe |
| DiGeorge Syndrome (22q11.2 Deletion Syndrome) | 22q11.2 (long arm of chromosome 22) | TBX1, COMT, DGCR8 | Heart defects, immune deficiency, cleft palate, developmental delays | Moderate to severe |
| Wolf-Hirschhorn Syndrome | 4p16.3 (short arm of chromosome 4) | WHSC1, LETM1, FGFR3 | Intellectual disability, seizures, distinctive facial features, growth delays | Severe |
| Smith-Magenis Syndrome | 17p11.2 (short arm of chromosome 17) | RAI1 | Intellectual disability, sleep disturbances, behavioral problems, distinctive facial features | Moderate to severe |
| 1p36 Deletion Syndrome | 1p36 (short arm of chromosome 1) | MMP23B, GABRD, SKI | Intellectual disability, seizures, heart defects, hypotonia | Moderate to severe |
| Miller-Dieker Syndrome | 17p13.3 (short arm of chromosome 17) | PAFAH1B1 (LIS1) | Severe brain malformations (lissencephaly), seizures, developmental delays | Severe |
| Jacobsen Syndrome | 11q23-q25 (long arm of chromosome 11) | FLI1, ETS1, NRGN | Intellectual disability, heart defects, bleeding disorders, distinctive facial features | Moderate to severe |
Duplications:
Duplication is an event in which a chromosomal part or region is duplicated. This means that it duplicates the gene copies covered during the duplication and increases the gene copies.
So,
When extra copies of chromosomal regions are formed, which results in copy number variation of genes within that particular area of the chromosome, it is known as duplication.
These duplicated regions lead to the overexpression of genes, which is responsible for several genetic abnormalities.
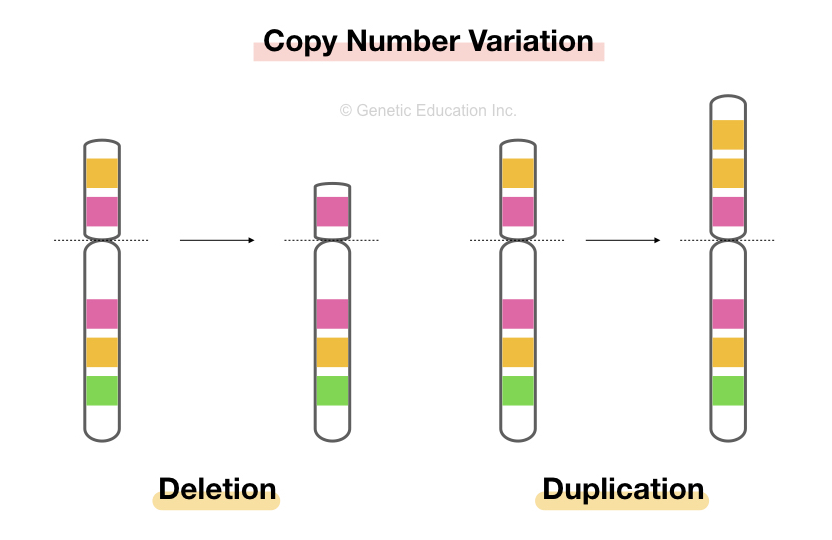
Mechanisms of Duplication:
The mechanism of duplication is similar to deletions, which arise due to unequal crossing over during meiosis, where homologous chromosomes misalign and as a result one chromosome gains an extra copy of a particular segment, meanwhile the other loses it. Duplications can also occur due to errors in DNA replication or repair.
Consequences of Duplications:
Again, the size and location of the duplicated segment are key factors here.
Duplications of the segments of a chromosome, which involve multiple genes, result in overexpression of those genes, leading to severe genetic disorders. Meanwhile, duplications of non-coding regions may remain asymptomatic.
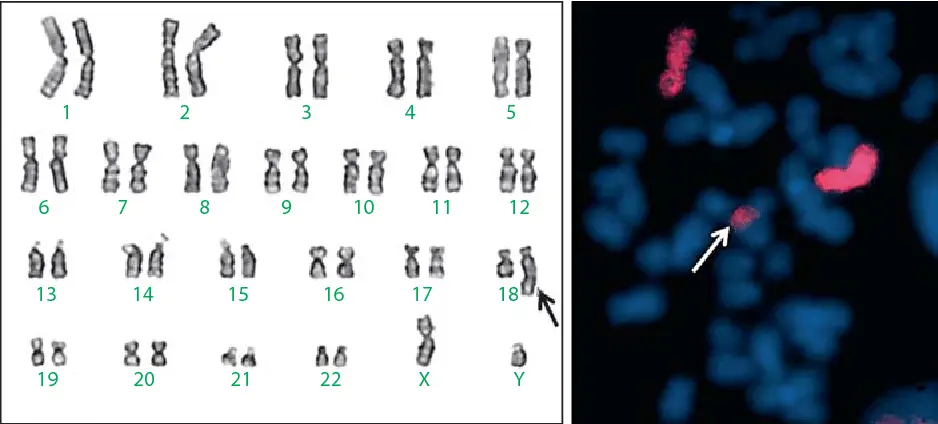
Examples of structural aberrations due to duplications, including chromosomes involved, genes involved, symptoms, and severity, are listed in the table below:
| Name of Condition | Chromosome Involved | Genes Involved | Symptoms | Severity |
| Charcot-Marie-Tooth Disease Type 1A | 17p12 (short arm of chromosome 17) | PMP22 | Peripheral neuropathy, muscle weakness, foot deformities | Moderate to severe |
| MECP2 Duplication Syndrome | Xq28 (long arm of X chromosome) | MECP2 | Intellectual disability, developmental delays, seizures, hypotonia | Severe |
| Pallister-Killian Syndrome | 12p (short arm of chromosome 12) | Multiple genes | Intellectual disability, distinctive facial features, seizures, hypotonia | Severe |
| Hereditary Neuropathy with Liability to Pressure Palsies (HNPP) | 17p12 (short arm of chromosome 17) | PMP22 | Recurrent nerve palsies, muscle weakness, sensory loss | Mild to moderate |
| Potocki-Lupski Syndrome (PTLS) | 17p11.2 (short arm of chromosome 17) | RAI1 | Intellectual disability, developmental delays, autism spectrum disorder | Moderate to severe |
| 16p11.2 Duplication Syndrome | 16p11.2 (short arm of chromosome 16) | Multiple genes | Intellectual disability, autism spectrum disorder, obesity, developmental delays | Mild to severe |
| 15q11-q13 Duplication Syndrome | 15q11-q13 (long arm of chromosome 15) | UBE3A, GABRB3 | Intellectual disability, autism spectrum disorder, seizures | Moderate to severe |
| 22q11.2 Duplication Syndrome | 22q11.2 (long arm of chromosome 22) | Multiple genes | Developmental delays, mild intellectual disability, speech delays | Mild to moderate |
| 1q21.1 Duplication Syndrome | 1q21.1 (long arm of chromosome 1) | Multiple genes | Intellectual disability, autism spectrum disorder, congenital anomalies | Mild to moderate |
| Xq28 Duplication Syndrome | Xq28 (long arm of X chromosome) | MECP2 | Intellectual disability, developmental delays, seizures, hypotonia | Severe |
Inversions:
When a segment of the chromosome breaks and reattaches in reverse orientation within the same chromosome, it is called chromosome inversion.
Inversions are of two types: paracentric inversion (doesn’t involve the centromere) and pericentric inversion (involves the centromere).
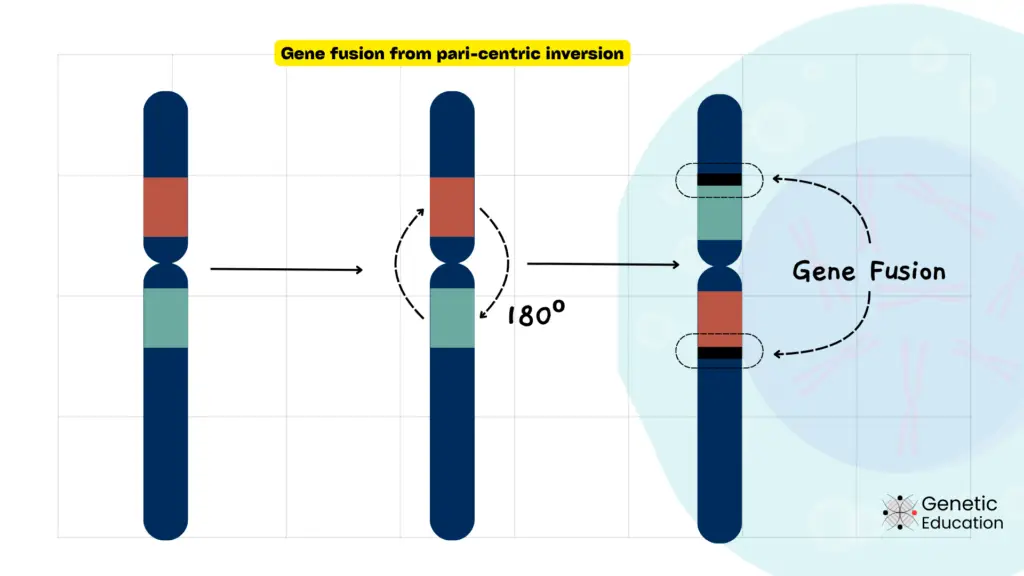
Mechanisms of Inversion:
During recombination or DNA repair, a segment of a chromosome breaks and is reinserted in a reversed orientation, resulting in chromosomal inversions. This can also happen as a result of exposure to mutagenic agents.
Consequences of Inversions:
When inversion of important genes or regulatory regions happens, it results in severe consequences leading to several genetic aberrations. While inversions in non-coding regions mainly remain asymptomatic.
Examples of structural aberrations due to inversions, including chromosomes involved, genes involved, symptoms, and severity, are listed in the table below:
| Name of Condition | Chromosome Involved | Gene Involved | Symptoms | Severity |
| Hemophilia A | Xq28 (long arm of X chromosome) | F8 | Bleeding episodes, joint damage, prolonged bleeding after injury or surgery | Moderate to severe |
| Inversion 9 | 9p11-q13 (chromosome 9) | None (pericentric inversion) | Often asymptomatic, but may be associated with infertility or miscarriage | Usually benign, but variable |
| FG Syndrome | Xq13 (long arm of X chromosome) | MED12 | Intellectual disability, hypotonia, distinctive facial features, constipation | Moderate to severe |
| Chromosome 3 Inversion | 3p14-q21 (chromosome 3) | None (pericentric inversion) | Often asymptomatic, but may be associated with developmental delays | Usually benign, but variable |
| Chromosome 10 Inversion | 10q11-q22 (chromosome 10) | None (pericentric inversion) | Often asymptomatic, but may be associated with infertility or miscarriage | Usually benign, but variable |
| Chromosome 16 Inversion | 16p11.2-q12.1 (chromosome 16) | None (pericentric inversion) | Often asymptomatic, but may be associated with developmental delays | Usually benign, but variable |
| Chromosome 18 Inversion | 18p11-q21 (chromosome 18) | None (pericentric inversion) | Often asymptomatic, but may be associated with developmental delays | Usually benign, but variable |
| Chromosome 4 Inversion | 4p15-p16 (chromosome 4) | None (paracentric inversion) | Often asymptomatic, but may be associated with developmental delays | Usually benign, but variable |
| Chromosome 8 Inversion | 8p23-q22 (chromosome 8) | None (pericentric inversion) | Often asymptomatic, but may be associated with developmental delays | Usually benign, but variable |
| Chromosome 11 Inversion | 11q13-q23 (chromosome 11) | None (pericentric inversion) | Often asymptomatic, but may be associated with developmental delays | Usually benign, but variable |
Ring Chromosomes:
A ring chromosome is formed when the chromosomal ends break and fuse together. This ring-like structure results in the loss of genetic material at the ends of the chromosome.
Mechanisms of Ring Chromosome Formation:
There are many mechanisms reported contributing to the ring chromosome formation. This mainly involves the telomere— the end and protecting chromosome caps.
Telomere dysfunction leads to the loss of telomeric sequences and induces end-to-end fusion. This happens by replication errors and exposure to mutagenic agents.
Chromosomal instability has also been involved in the ring chromosome formation, research showed.
In addition, double-stranded DNA breaks on chromosome arms, and if it loses telomeres, also leads to ring chromosome formation.
Consequences of Ring Chromosomes:
Ring chromosome formation causes loss of genetic material and gene fusion. Loss of genetic material has been significantly reported, though.
Thus, the severity of the abnormal condition governed by the ring chromosome formation depends on the amount of genetic material lost during the event.
Examples of structural aberrations due to the formation of a Ring chromosome, including chromosomes involved, symptoms, and severity, are listed in the table below:
| Name of Condition | Chromosome Involved | Symptoms | Severity |
| Ring Chromosome 20 Syndrome | 20 | Epilepsy, intellectual disability, behavioral problems | Moderate to severe |
| Ring Chromosome 14 Syndrome | 14 | Intellectual disability, seizures, distinctive facial features | Moderate to severe |
| Ring Chromosome 13 Syndrome | 13 | Intellectual disability, growth delays, microcephaly, heart defects | Severe |
| Ring Chromosome 18 Syndrome | 18 | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Ring Chromosome 15 Syndrome | 15 | Intellectual disability, seizures, developmental delays | Moderate to severe |
| Ring Chromosome 22 Syndrome | 22 | Intellectual disability, developmental delays, hypotonia | Moderate to severe |
| Ring Chromosome 9 Syndrome | 9 | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Ring Chromosome 6 Syndrome | 6 | Intellectual disability, growth delays, microcephaly | Moderate to severe |
| Ring Chromosome 17 Syndrome | 17 | Intellectual disability, developmental delays, seizures | Moderate to severe |
| Ring Chromosome 4 Syndrome | 4 | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
Isochromosome:
A structural aberration is when one arm of the chromosome is duplicated while the other one is deleted, resulting in two identical arms which are mirror images of each other. This is an unbalanced structural aberration, most commonly seen in the X chromosome.
Mechanisms of Isochromosome Formation:
During meiosis, in normal conditions, chromosomes divide longitudinally, but in some cases, chromosomes divide transversely, which results in two identical arms, one from each sister chromatid. This misdivision of centromere results in Isochromosomes.
Consequences of Isochromosomes:
During this misdivision of centromere, one arm gets duplicated while the other arm gets deleted. If the arm being lost contains genetic information, which is important for the normal development and functioning of the cells it produces, a genetic abnormality.
Examples of structural aberrations due to the formation of Isochromosomes, including chromosomes involved, symptoms, and severity, are listed in the table below
| Name of Condition | Chromosome Involved | Genes Involved | Symptoms | Severity |
| Turner Syndrome (Isochromosome Xq) | X | SHOX, RPS4X | Short stature, ovarian failure, cardiovascular abnormalities | Moderate to severe |
| Pallister-Killian Syndrome | 12p (short arm of chromosome 12) | Multiple genes | Intellectual disability, distinctive facial features, seizures, hypotonia | Severe |
| Isochromosome 18p Syndrome | 18p (short arm of chromosome 18) | None (loss of long arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Isochromosome 9p Syndrome | 9p (short arm of chromosome 9) | None (loss of long arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Isochromosome 8p Syndrome | 8p (short arm of chromosome 8) | None (loss of long arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Isochromosome 5p Syndrome | 5p (short arm of chromosome 5) | None (loss of long arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Isochromosome 17p Syndrome | 17p (short arm of chromosome 17) | None (loss of long arm genes) | Intellectual disability, developmental delays, seizures | Moderate to severe |
| Isochromosome 21q Syndrome | 21q (long arm of chromosome 21) | None (loss of short arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Isochromosome 7q Syndrome | 7q (long arm of chromosome 7) | None (loss of short arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
| Isochromosome 10q Syndrome | 10q (long arm of chromosome 10) | None (loss of short arm genes) | Intellectual disability, growth delays, distinctive facial features | Moderate to severe |
Wrapping up:
Structural chromosomal anomalies have been commonly reported in cancer cases; however, it is also involved in other genetic abnormalities as well. With translocations being the most common, other anomalies like duplication, deletion or inversion are also reported.
Common causes that lead to such structural chromosomal aberrations are either intrinsic factors— replication errors, DNA repair failure or cell cycle faults or extrinsic factors— exposure to radiation, mutants or chemicals.
Techniques like karyotyping, FISH and chromosomal microarrays are extensively employed to study structural chromosomal anomalies. Karyotyping is the gold-standard method, though.
I hope you like this article and the information we collected. Share it with your friends and subscribe to Genetic Education.
References:
Antonarakis, S. E., & Rossiter, J. P. (2001). Factor VIII gene inversions in severe hemophilia A. Blood, 97(1), 1-2. https://doi.org/10.1182/blood.V97.1.1
Antonarakis, S. E., Lyle, R., & Dermitzakis, E. T. (2004). Chromosome 21 and Down syndrome: From genomics to pathophysiology. Nature Reviews Genetics, 5(10), 725-738. https://doi.org/10.1038/nrg1448
Cassidy, S. B., & Driscoll, D. J. (2009). Prader-Willi syndrome. European Journal of Human Genetics, 17(1), 3-13. https://doi.org/10.1038/ejhg.2008.165
Deininger, M. W., Goldman, J. M., & Melo, J. V. (2000). The molecular biology of chronic myeloid leukemia. Blood, 96(10), 3343-3356. https://doi.org/10.1182/blood.V96.10.3343
Gardner, R. J. M., Sutherland, G. R., & Shaffer, L. G. (2012). Chromosome abnormalities and genetic counseling (4th ed.). Oxford University Press. https://doi.org/10.1093/med/9780195375336.001.0001
Griffiths, A. J. F., Wessler, S. R., Carroll, S. B., & Doebley, J. (2015). Introduction to genetic analysis (11th ed.). W.H. Freeman and Company.
Inoue, Y., Fujiwara, T., & Matsuda, K. (1997). Ring chromosome 20 and nonconvulsive status epilepticus. Epilepsia, 38(2), 210-213. https://doi.org/10.1111/j.1528-1157.1997.tb01100.x
National Human Genome Research Institute (NHGRI). (2023). Chromosome abnormalities. Retrieved from https://www.genome.gov
National Organization for Rare Disorders (NORD). (2023). Cri-du-chat syndrome. Retrieved from https://rarediseases.org
Nussbaum, R. L., McInnes, R. R., & Willard, H. F. (2016). Thompson & Thompson genetics in medicine (8th ed.). Elsevier.
Pareyson, D., & Marchesi, C. (2009). Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. The Lancet Neurology, 8(7), 654-667. https://doi.org/10.1016/S1474-4422(09)70110-3
Pober, B. R. (2010). Williams-Beuren syndrome. New England Journal of Medicine, 362(3), 239-252. https://doi.org/10.1056/NEJMra0903074
Ramocki, M. B., & Zoghbi, H. Y. (2008). Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature, 455(7215), 912-918. https://doi.org/10.1038/nature07457
Strachan, T., & Read, A. P. (2019). Human molecular genetics (5th ed.). Garland Science.
Sybert, V. P., & McCauley, E. (2004). Turner’s syndrome. New England Journal of Medicine, 351(12), 1227-1238. https://doi.org/10.1056/NEJMra030196
Teschler-Nicola, M., & Killian, W. (1981). Pallister-Killian syndrome: A review. American Journal of Medical Genetics, 39(3), 279-283. https://doi.org/10.1002/ajmg.1320390302
Zollino, M., et al. (2003). Ring chromosome 14 syndrome: A clinical and molecular study. American Journal of Medical Genetics, 117A(1), 1-8. https://doi.org/10.1002/ajmg.a.10879