EMSA stands for Electrophoretic Mobility Shift Assay often known as gel shift assay is a technique to study the nucleic acid and protein interaction. The present technique is used ‘in addition to’ DNA footprinting.
Affinity electrophoresis, gel retardation assay, gel mobility shift or band shift are several names used for the gel shift assay. As mentioned in our previous article, protein-nucleic acid interaction has pivotal importance to on/off gene regulation.
Moreover, in vitro studies on how ligands, receptors or antibodies interact with a sequence can be ruled out as well. Basically, it is a modification of conventional gel electrophoresis and relies on the migration of DNA under an electrical current.
The native electrophoresis either conventional agarose gel or polyacrylamide gel electrophoresis effectively separates biological molecules like protein or DNA; thereby determining the size. Although, in the current scenario, it has other lucrative applications too.
In the present article, I will explain to you the principle of the EMSA, how it works and its importance in the protein DNA interaction study.
Stay tuned.
Quick Summary:
| Name | Gel shift assay |
| Application | Protein-nucleic acid interaction studies |
| Other names | Gel mobility shift assay, EMSA, electrophoretic mobility shift assay, band shift assay and gel retardation assay. |
| Investigations | Qualitative |
| Native technique | SDS-PAGE |
| Advantage | Required small sample volume |
| Disadvantage | Inaccurate and provide limited data |
Key Topics:
What is gel shift assay or EMSA?
The gel shift assay is used with the DNase assay, footprint or primer extension assay to explicate the DNA/RNA-protein interactions. The present technique, used in the research is based on the technique described by Garner & Revzin, however, the assay was originally described by Fried and Crothers.
Put simply, gel shift assay is a qualitative assay, that results in visuals and is interpreted by manual evaluations. Henceforth, it comes with so many limitations.
Related article: What is DNA footprinting?- Principle, Steps, Process and Applications.
Principle:
The native principle of agarose gel electrophoresis has been utilized to study the interaction and chemistry between nucleic acid and various amino acid complexes. When the nucleic acid linked and unliked samples are loaded in a polyacrylamide gel and run under an electric field, with standard gel running conditions; a mobility shift is observed.
Depending upon the size of the sample (which is any protein bound to the DNA), differential migration speed will be seen, showing that the unlinked nucleic acid runs faster than the protein-linked one. The slower migration speed results in the “shift” of bands and differential banding patterns.
Technically, in a gel- well with the protein-nucleic acid bound sample, we will get two separate bands. Results help to investigate the part of nucleic acid to linkage with a protein and the structural stability, site of binding & the part of a protein-linked if the concentration is known.
The assay is often known as the gel retardation or shift assay as the binding affinity of protein retards or shifts the migration speed of target DNA.
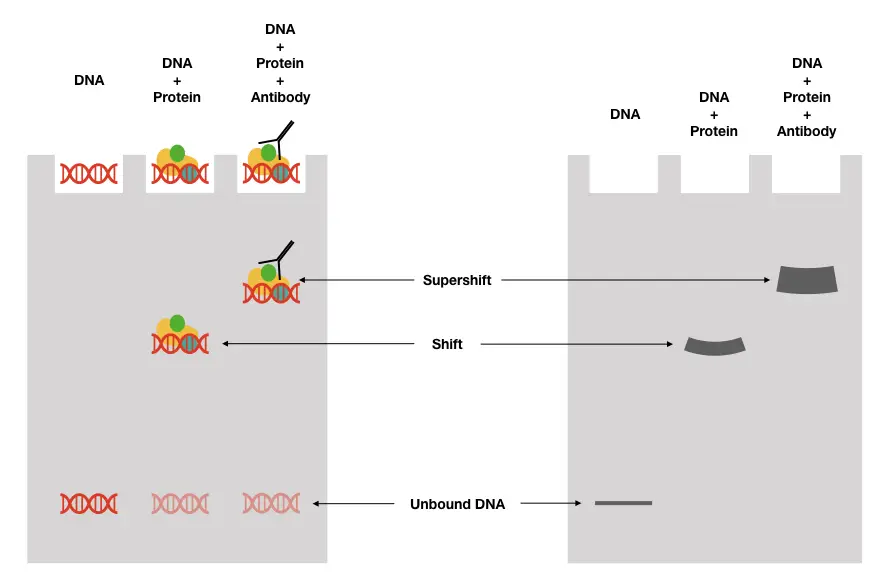
Process of gel shift assay:
The typical process of shift assay includes steps like selecting the target DNA or probe DNA, labeling the probe, competitive binding with the protein, gel electrophoresis and detection.
Selecting the target or probe DNA:
Generally, 100 to 500bp longer DNA sequencing should be selected with definite supercoiling. Fragments smaller than 100bp give non-specific results, in addition, circular DNA-linked protein complexes migrate faster and give non-conclusive results.
If longer sequences are used and digested for further investigations before the gel shift assay, the restriction enzyme is removed by purification first. Sequences that form a secondary structure and have repetitive DNA are usually not selected or purified by high-performance liquid chromatography, before use.
It is also important to note that the sequence or template used in the assay should be known or sequenced. Less concentration of the targeting material is used to avoid non-specific binding during the reaction.
Gel preparation:
An important step the present technique has is gel preparation. The gel must be polyacrylamide as it has higher resolution power and should be a non-denaturing gel assay to prevent dissociation of the complex.
The gel and other chemicals are prepared using a standard gel preparation protocol. Moreover, it is also important to set gelling codings in such a way that it should not affect the structure, stability and migration rate of protein-DNA complexes.
Labeling or staining:
Two common techniques to study the results of the PAGE gel shift assay are gel staining and nucleic acid labeling. EtBr (Ethidium Bromide) is a popular base stacking fluorochrome used to visualize the DNA. Here the gel is stained with the EtBr overnight, and under the illumination, it gives a differential banding pattern.
Nonetheless, the present technique is too poor and produces strong background sounds by staining the part of a gel too. So not advised.
Yet another and most popular method is DNA labeling either by radio-labeling or fluoro labeling. We already have discussed both types of nucleic acid labeling in our previous article, you can read there.
In short, 32P as a radiolabel or SYBR green dye as fluoro-labeled is incorporated at either end of the DNA and allowed to autoradiography and illumination, respectively to get results. Note that each technique has its own advantages and limitations.
Advantages:
The present technique is simple, reliable, speedy, accurate and more effective.
It needs a smaller starting volume >20microliter and a separate total protein or DNA concentration up to 0.1nM.
Limitations:
It can provide qualitative results only.
Adverse gelling conditions may result in abnormal or retarded results by migration shift.
Impure crude cell extract and a highly pure protein both can be used in the present assay.
Applications:
Hellman & Fried, 2007 explained some of the excellent variants by that applications of gel shift assay in their paper entitled, “Electrophoretic mobility shift assay (EMSA) for the detecting protein-nucleic acid interactions.”
The present technique is employed to study and measure association and dissociation kinetics of protein and DNA, and determine the binding stoichiometry using assays like double layer and continuous EMSA.
Techniques like circular permutation and phased bands analysis with the EMSA have fruitful utilization for detecting DNA bending like protein-induced DNA bending.
Moreover, standard optimizations like reverse EMSA and binding partition analysis have been used for measuring protein binding as well as nucleic acid binding affinity; nucleic acid binding affinity with multiple proteins and a single protein binding affinity for various nucleic acid targets.
When a topoisomerase enzyme is used in the native EMSA technique, it can be utilized as a tool for investigating the protein and supercoiled DNA complex, corresponding to protein-DNA interactions within the nucleosome complex and can be determined by nucleosome shift assay.
Kristie & Roizman, 1986 explained the antibody binding assay using the native gel shift assay which studies a protein with a specific epitope and how it affects the mobility of the protein-DNA complex.
Bain & Ackers, 1998 elucidated a gel shift assay named “quantitative cryogenic gel shift assay” for detecting labile complexes.
Other techniques like protein-protein, DNA-DNA and RNA-RNA gel retardation are used to study protein-protein, DNA-DNA and RNA-RNA interactions in the absence of DNA/RNA, protein and other protein complexes, respectively.
Besides enlisted concrete applications, the present technique even becomes more aggressive when combined with other state-of-the-art methods. EMSA along with MS, for example, is used to identify unknown binding proteins to the nucleic acid sequence.
Likewise, the combination of EMSA, SDS-PAGE and western blot has been used for the same purpose as explained.
Trending optimizations, advancements and variations in EMSA:
EMSA includes many variants, scientists use as per the requirement of the assay or to elucidate different purposes. In this section of the article, I will explain some trending, useful and optimized EMSA variants. This section is especially useful for research scholars.
EMSA Supershift:
‘Supershift ‘ means slowing down the migration process even at a reduced rate due to the presence of another factor, molecule or protein. This has importance in studying crude cell extract which is an impure suspension of various proteins and nucleic acids.
For instance, the binding of antibodies to the transcriptional factor -DNA complex. This larger complex induces “supershift”, which is, when compared with a conventional shift assay or complex lacking a specific antibody, the “super-complex” can be studied.
What the results portray is how the binding of TF to DNA, along with other factors or proteins is used by a cell for various processes. Notedly, only a nondenaturing gel assay is used here.
2D-EMSA:
Yang et al., in their article, “synthetic biology and metabolic engineering in plants and microbes part B: Metabolism in plants.” published in 2016 explained important modification of 2D-EMSA.
When combined with Mass spectroscopy like the MS-MALDI-TOF; the target protein extracted from bacterial lysate or crude can be precisely identified. As aforementioned in the supershift assay binding site for a specific antibody, an antibody linked to the transcriptional factor and conformational changes by binding; all it can investigate.
However, it works only when the binding factor (transcriptional) will be known and intentionally used. Nonetheless, low-abundance transcriptional factors and complex nuclear crude extract can’t be studied well.
ChIP-seq is a powerful, state-of-the-art assay and a quantitative method to study the protein DNA interaction as well as the sequence involved in the same at a chromatin level.
You can read more on this technique here: The Concept of ChIP-Seq (ChIP-Sequencing) Explained.
Wrapping up:
Together with the DNA footprint assay, the gel shift technique or EMSA allows determining the protein-DNA interactions more clearly, precisely and quantitatively, when combined with other techniques.
The technique is one of the important tools for transcriptional, transcriptional factor activity and TF-DNA interaction studies. Along with this, it also determines how various TF-DNA complex with other proteins performs various functions in various cells.
I hope this article makes sense and helps you to understand the importance of epigenetic interactions.
Sources:
Hellman, L. M., & Fried, M. G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nature protocols, 2(8), 1849–1861. https://doi.org/10.1038/nprot.2007.249.
Jett, S. D., & Bear, D. G. (1994). Snapshot blotting: transfer of nucleic acids and nucleoprotein complexes from electrophoresis gels to grids for electron microscopy. Proceedings of the National Academy of Sciences of the United States of America, 91(15), 6870–6874. https://doi.org/10.1073/pnas.91.15.6870.
Kristie, T. M., & Roizman, B. (1986). Alpha 4, the major regulatory protein of herpes simplex virus type 1, is stably and specifically associated with promoter-regulatory domains of alpha genes and of selected other viral genes. Proceedings of the National Academy of Sciences of the United States of America, 83(10), 3218–3222. https://doi.org/10.1073/pnas.83.10.3218.
Bain, D. L., & Ackers, G. K. (1998). A quantitative cryogenic gel-shift technique for analysis of protein-DNA binding. Analytical Biochemistry, 258(2), 240–245. https://doi.org/10.1006/abio.1998.2626.
Yang et al., Chapter eleven- Establishing the Architecture of Plant Gene Regulatory Networks. Methods in Enzymology, 576, 251-304.

