“An in silico PCR is a computational tool used to determine the results of practical PCR reaction computationally using a set of the selected primers.”
Here in the in silico PCR, we are performing a PCR reaction virtually, on computer software, therefore it is also known as electronic PCR or virtual PCR. The objective of performing this type of computational analysis is to predict the result of actual PCR reaction.
But why to do in silico PCR? because, we can get some rough idea of how our reaction behaves during the actual PCR. Thus if it is not good, it saves time, chemicals, buffer and man-power.
Actually it is nota true PCR reaction, thus dNTPs, primers, buffer and DNA are not used in it. Instead, only sequence information and primer data is needed to do analysis.
To understand the concept of the in silico PCR, we have to answer this question,
Why the in silico PCR is required?
For that, we have to first understand the meaning of the in silico and
- What is in silico?
- What is in silico PCR?
- What is a primer and what type of primers are used in the PCR?
Read our interesting article on PCR: A Complete Guide of the Polymerase Chain Reaction.
What is in silico?
Wikipedia defines the term in silico as “perform on a computer or via computer simulation.”
“Any process done on the computer-based application is called in silico process.”
Different types of in silico PCR software are available online. Some of them are free some are paid. However, the most popular software is ePCR and in silico PCR by the UCSC genome browser.
In this article, we are giving you a comprehensive guide on how to do in silico PCR.
So let’s start.
First of all, I will explain to you in silico PCR in a simple language,
Key Topics:
What is in silico PCR?
In silico PCR is a computer application based analysis of our PCR reaction with the help of the primer information. Here, the software checks the primers (what we designed) against the genome of our interest.
It actually, examines the primer set that we selected against the target sequence or genome. It gives the idea of at which position the primer going to binds, whether it is bind to the exact target location or any other non-specific location.
The software also interprets the results computationally and gives a rough idea about the product size of the amplicon. By doing this we can analyze the efficiency and the sensitivity of the primers.
Now lets roughly understand about the primers.
What is the primer?
The primers are the short single-stranded DNA oligonucleotides used to amplify DNA in vitro. It provides the 3′ end to the Taq DNA polymerase to expand the growing DNA strand.
The primer must be 18 to 22 nucleotides long, having 50% of GC content and can bind only at the specific location to our DNA of interest. For more detail on PCR primer please read our article: PCR primer design guidelines
The prime is a very important component of the PCR reaction. We have to design primers every time for every different PCR reaction. For different PCR template DNA, primers are different.
One of the important properties of any primer is its specificity.
The primer must be specific to one target. If it binds to more than one location, it will give non-specific bands of the DNA, non-specific binding is the major limitation of any primer sequences.
The in silico PCR facilitates the computational analysis of our PCR reaction. Just like the primer 3, it will give a brief idea about the nature of the primer, how it binds, where it binds and what results, it will give at the end.
Read more,
In silico PCR provides information about orientation, binding site, location, amplicon length and open reading frame identification of primer by using the whole genome or mRNA data of the organism.
Now take a look at the quick guide on How to do in silico PCR:
I am explaining to you the most common and widely used in silico PCR by the UCSC genome browser. The link is here the UCSC genome browser.
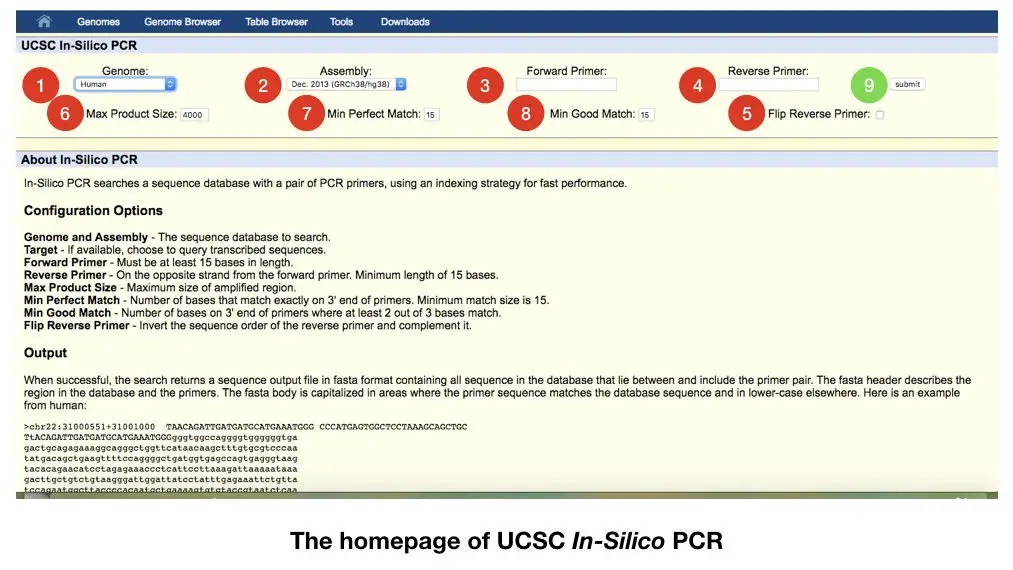
The figure above is the home page on the UCSC genome browser. Once you open the link, you will land on this page.
Now, we have to customize several settings lets start with the genome.
See the red labeled step 1: choose the genome, if you are working on any plant species select it. If you are working on human DNA select the human genome. We have selected here the human genome.
Step 2: select the assembly. Here we have to choose which available assembly we want to use, generally, we are using Dec.2013(GRCh38/gh38).
Read more:
After that in step 3 paste our forward primer in the 3rd box and reverse primer in the 4th box. The primer can be designed by using the primer 3 software. Read how to design primers in primer 3: PCR primer design guidelines
After that in the 5th step select flip primer if you want to flip it or retain it as well. It is up to us, what type of reverse primer we have.
Now in step 6 select the maximum product size. By default it is 4000, we can customize it if required. As per the requirement, we can set it 1000bp, 2000bp of 3000bp. Our product must remain in the range we selected.
In step 7 select the perfect match, we can customize it one or two, However, if our primers are perfect to our target sequence, we will get only a single product.
I personally select it by default, why?
Because the result gives us an idea about how many positions into the genome our primer can bind so that we can encounter the non-specific bindings.
In step 8 select the number of how many good matches we want.
After customizing all the steps, submit the data in step 9. The result of the an in silico PCR is as shown into the figure below,
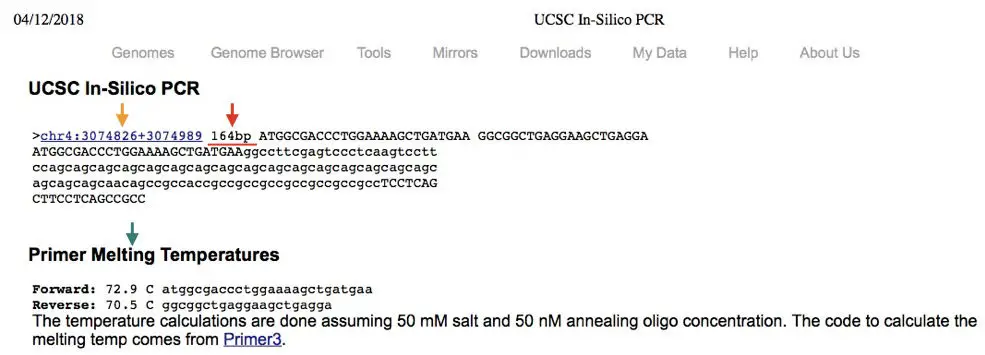
Now, what we will get into the results?
See the image, the Yellow arrow, it is the location of the forward and reverse primer binding site into the genome that we had selected.
Our primer binds on chromosome number 4, between the position 3074826 to 3074989.
Next, see the red arrow. What we get after the in silico PCR is the 164 bp PCR product.
Now, the green arrow indicates the annealing temperature and other information of the primers.
After doing in silico PCR we can interpret that, our primers are specific to our target sequence only, which is located on chromosome number 4 and the PCR product will be 164 bp long.
Therefore, when we will perform the actual PCR reaction, our product after agarose gel electrophoresis must be 164 bp. If it will not then our product is non-specific or the result is false.
Furthermore, cross-check the in silico PCR results with the primer 3.
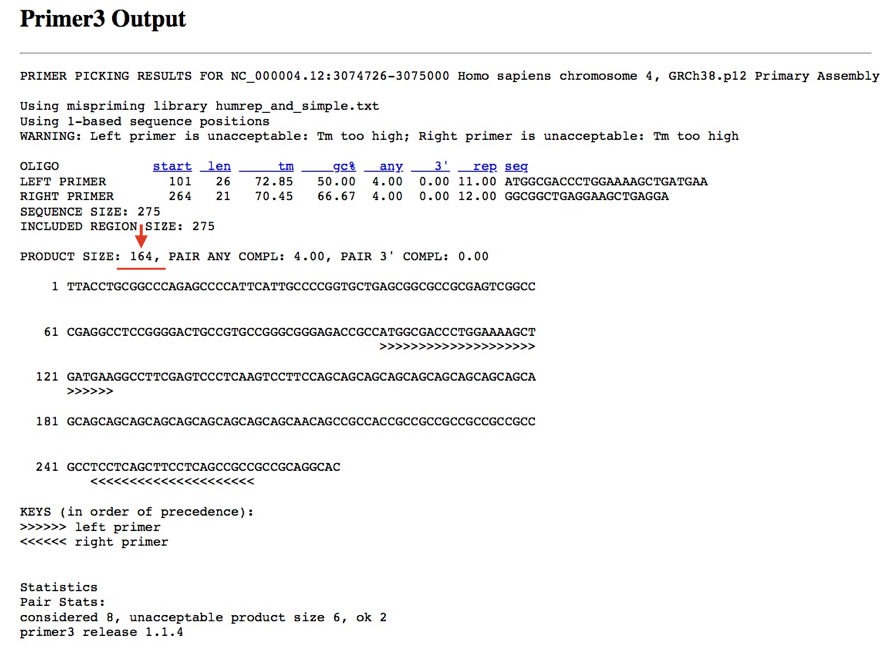
See the results, our product size, 164bp. Our primer is specific to the target DNA of our interest.
The final results of the UCSC genome browser are like this,
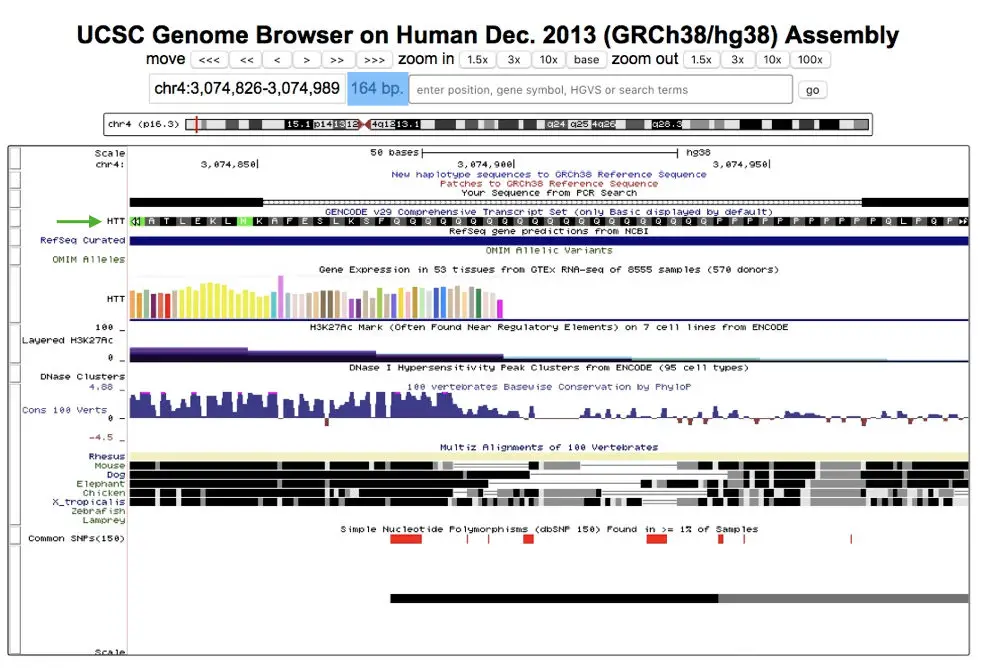
See the green arrow, our gene is on chromosome 4 and it is Huntington’s gene (HTT). Other information related to the target sequence is there into the window, analyze it by doing it yourself:
[epcl_button label=”In silico PCR” url=”http://genome.ucsc.edu/cgi-bin/hgPcr?db=hg18″ type=”flat” color=”dark” size=”regular” icon=”fa-check” target=”_blank”][/epcl_button]
Other in silico PCR are,
In silico PCR for different bacterial genome click here.
Advantages of in silico PCR:
We can amplify (e-amplify) the sequence against any genome or transcriptome.
It actually optimized our primers and increases the specificity and the sensitivity of the reaction.
All the available software almost predicts accurate results.
Also, the in silico PCR saves money and time by predicting the results before doing the PCR.
Conclusion:
In silico, PCR is a good tool to predict the outcome of the reaction. Everyone should learn it. Along with the wet lab work, dry lab work is important as well. Other bioinformatics tools such as BLAST, PRIMER-BLAST and eSTS-PCR are also more useful. We will discuss is in some other articles.



Very precise and nice explanation. However the universal web facility like NCBI could have been also included for it will offer web facility for plant genomes also so that the illustration could have been complete–covering animals, plants and microorganisms.