In 1963, J Cairns reported the process of replication in E.coli bacteria by autoradiography.
Replication is a process in which a DNA molecule is copied. From the double-stranded DNA, two daughter dsDNA is generated and is transferred to two different daughter cells.
Before proceeding to this article please read the General process of DNA replication. It will give you a brief idea about replication. The process of prokaryotic replication is divided into three broad categories:
- Initiation
- Elongation
- Termination
Key Topics:
Initiation of Replication
In E.coli the process of replication is initiated from the origin of replication. The origin of replication in E.coli is called as oriC.
oriC consists of a 245bp long AT-rich sequence which is highly conserved in almost all prokaryotes. Mostly two types of sequences are present in this region, three repeats of 13bp called as a 13mer and five repeats of 9bp called as a 9mer.
A few proteins play an important role in DNA replication:
- DnaA– it recognizes oriC sequences for initiation of replication.
- DanB– unwinds DNA. It is actually a helicase.
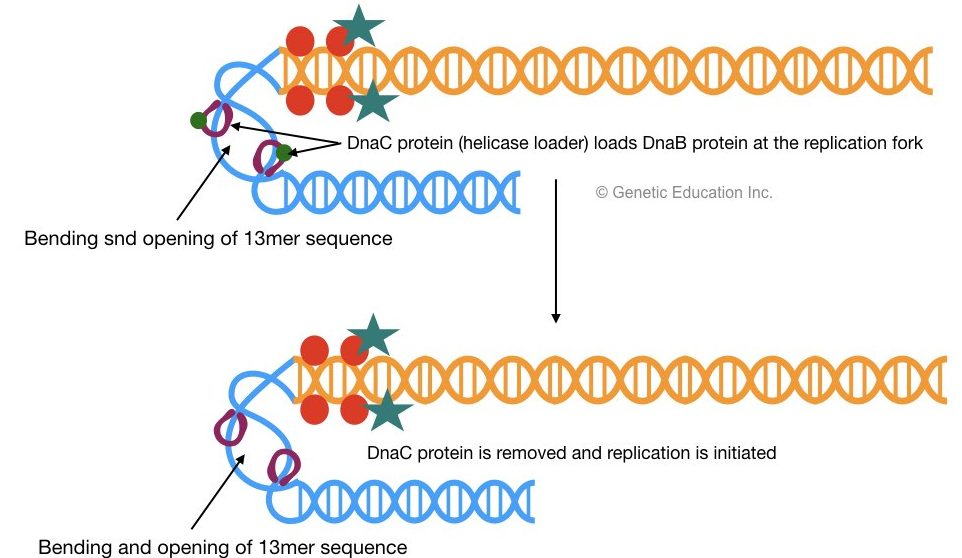
- DnaC– at the origin of replication it helps helicase (DnaB) to recognize the site for its action.
- DnaG– it is actually a primase. It synthesizes a new RNA primer.
- DNA gyrase– it is a DNA topoisomerase II which helps in the unwinding of DNA.
A higher concentration of protein A signals to start cell division. Protein A (Dna A protein) with ATP, binds to the 9mer sequences of oriC. This binding allows the opening of 13mer sequences of oriC by bending the DNA.
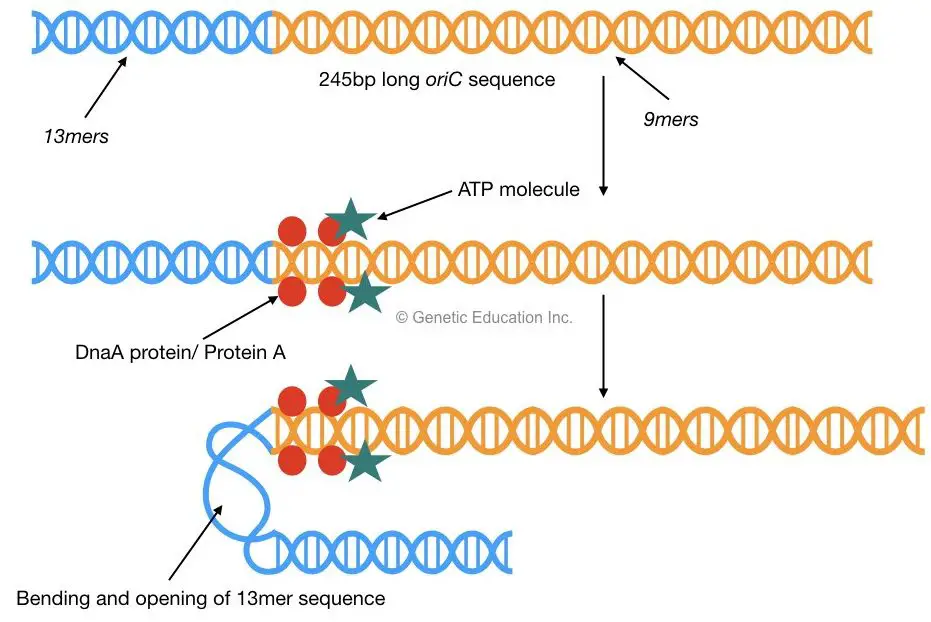
2 DnaB proteins bind to 13mer repeats which are recognized by DnaC. Once DnaB is settled on oriC (at 13mer) the DnaC protein will be released.
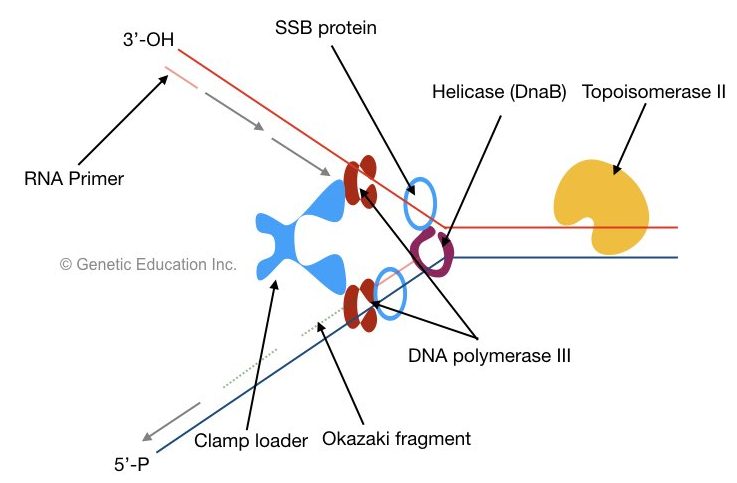
DnaB helps in the unwinding of DNA and generates two single-stranded DNA molecules, one with a 5’-P end and another with the 3’-OH end. Helicase uses ATP as an energy source for the unwinding of DNA.
Read more on Helicase: What Is Helicase? And How It unwinds DNA?
The activity of helicase (Dna B) generates tension on the remaining double strand of DNA. DNA gyrase helps in the unwinding of DNA and releases the tension on the DNA strand by negative supercoiling. Tensioned or supercoiled DNA decreases the rate of replication.
For more detail on gyrase read the article: DNA Topology class 1: Topoisomerase
Here as the oriC is recognized, the opening of the DNA strand creates a replication fork. The replication fork is a Y-shaped structure and it is important because it facilitates polymerase to work properly.
As the chromosome (so-called chromosome) of bacteria is circular, two different replication forks are developed in two different directions and it runs towards each other.
When helicase unwinds DNA, SSB proteins bind the single strand near helicase. The single-stranded binding protein binds to the single strand of DNA and protects it from rejoining.
SSB is a tetrameric structure that protects the DNA and facilitates helicase activity constantly.
Read more on SSB protein: Single-Stranded Binding Protein: Structure And Function
It also prevents single-stranded DNA from the attack of nuclease. As the helicase Starts unwinding, polymerase starts adding nucleotide in the elongation stage.
Elongation of Replication
Before the start of elongation, primase synthesized a short 10bp long RNA primer. The DNA polymerase can not able to add nucleotide without an RNA primer. It provides a start site as a free 3’-OH group for the polymerase to work.
Read more on RNA primer: Meet DNA Primase: The Initiator Of DNA Replication
The elongation is a step in which the DNA synthesis is initiated. once the dsDNA becomes single-stranded, the polymerase settles on the junction of the DNA-RNA primer.
The first DNA polymerase is isolated by Arthur Kornberg, in 1959 and he was the first person who synthesized the first DNA molecule in vitro. Three different types of DNA polymerase helps in prokaryotic DNA replication.
DNA polymerase I: removes RNA primer by exonuclease activity and proofreads the DNA
DNA polymerase II: DNA repair function. DNA polymerase II mainly involved in the DNA repair pathway.
DNA polymerase III: adds nucleotide from 5’ to 3’ direction.
Once a primosome complex is created (primosome complex is a primer – Helicase complex) polymerase recognized it and starts the polymerization process at the leading strand.
The process of replication progressed from the 3’ to 5’ direction while the polymerase III adds nucleotide from 5’ to 3’ direction.
Both sentences are confusing, right? the figure shows how it is possible:
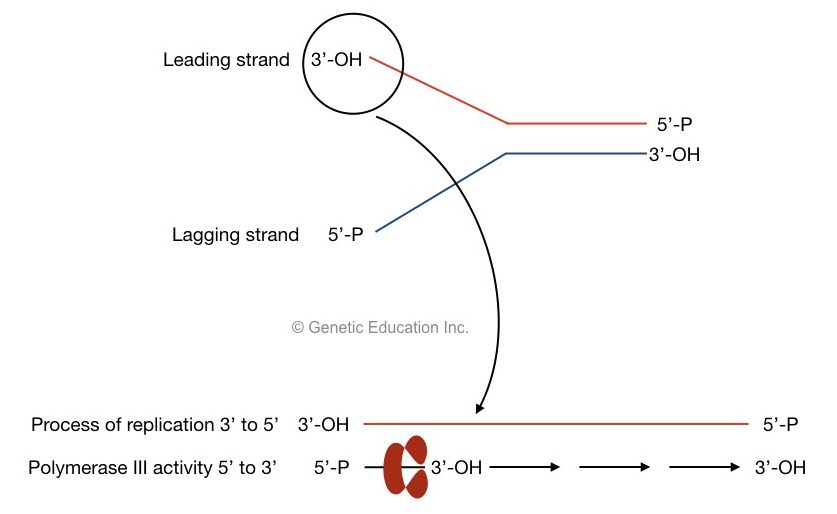
The activity of DNA polymerase:
Two DNA polymerase synthesized both leading strands as well as lagging stand at once. It is possible with the help of two subunits of DNA polymerase III.
Both subunits have different properties as one is specific for leading strand synthesis and one is specific for lagging strand synthesis.
Two sliding clams and a polymerase clam loader help polymerase to settle on each DNA strand. The clamp loader places the sliding clamp onto the DNA. The sliding clam holds DNA properly and prevents them from floating off.
Read our amazing article on DNA: DNA story: The structure and function of DNA
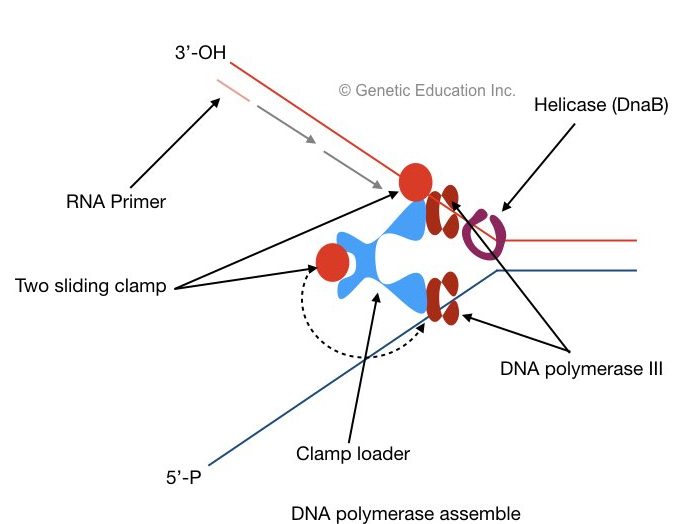
Leading strand synthesis:
A 3’- OH end of single-stranded DNA is called a leading stand. Primer is first bound to this end, after the recognization of the primer binding site, as the helicase moves, polymerase III adds nucleotides from 5’ to 3’ direction on growing leading strand.
During the process of replication 1000 nucleotides are added per second.
The leading strand DNA synthesis is very straightforward. It starts at the primosome and continuously adds nucleotides until the termination sequences.
Termination sequences are unique conserved sequences that are recognized by polymerase as the end of replication.
Lagging strand DNA synthesis:
The lagging strand of DNA has the 5’-P open end, hence the polymerase cannot synthesize new DNA from this direction.
The lagging strand DNA synthesis is a little different from the leading strand. Here primase synthesizes multiple primers.
Attend the class: DNA topology
the lagging strand is synthesized in short DNA fragments called Okazaki fragments.
Additionally, SSBP helps the single-stranded DNA from the attack of nuclease. It is present throughout the replication process to protect single-stranded DNA.
Subsequently, each new primer is added after 1000 nucleotides. After the synthesis of the Okazaki fragment, the sliding clamp is removed.
Once again, the new primer is synthesized by primase and binds to DNA, the clamp is further loaded by sliding clamp loader and the synthesis of the new Okazaki fragment begins.
Termination of Replication
After the synthesis of the leading and lagging strand, the polymerase is detached from the site of replication.
In the termination step, firstly, the multiple primers at the lagging strand are cleaved by RNase H and removed by DNA polymerase I. It also fills the gap between two Okazaki fragments by the addition of nucleotides.
Furthermore, DNA polymerase proofreads the sequence for avoiding errors in replication.
Finally, the enzyme DNA ligase fills the gap (creates a phosphodiester bond between Okazaki fragments and newly added nucleotides).
Read our article on DNA ligase: What Is DNA Ligase? And How T4 DNA Ligase Works?
At the last stage of termination, two replication fork meets at terminator recognizing sequences, called a Ter.
Ter sequences with TUS protein create a complex which arrests the replication fork and prevents them to escape.
At this complex, the process of replication is completed and all other proteins and enzymes leave this site.
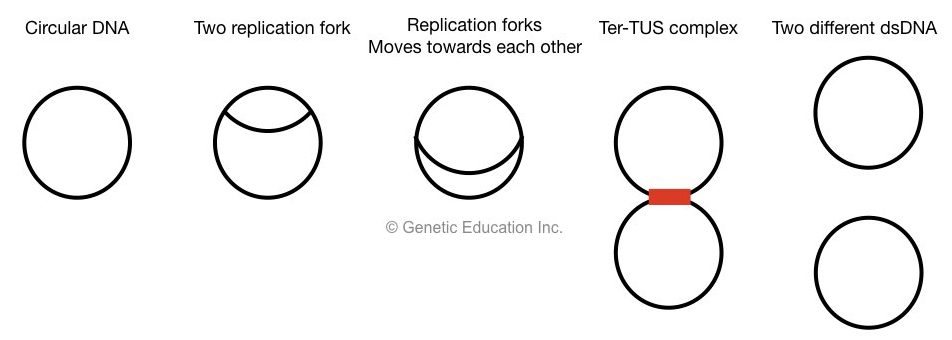
Only DNA topoisomerase II remains in action, it cuts both strands, dissociates Ter-TUS complex and two different circular DNA is generated.
Evidence of replication:
J Cairns first observed replication in E.coli bacteria by autoradiography.
He grew E.coli cells in a medium containing 3H-thymidine and collects the chromosome by lysing the cell at various time intervals. He transferred the chromosome to the filter membrane without breaking the chromosome.
The photographic film was developed and the result was obtained as mentioned:
The E.coli chromosome is circular in nature.
During the new strand synthesis, it exists a θ shaped structure which indicates that replication is initiated from two points in a circular chromosome.
He named the start point the origin of replication. The process of prokaryotic replication is semiconservative.
The two Y-shaped structure is actually replication fork that runs toward each other.
However, he postulated some of the processes incorrectly, as he indicated that the process is unidirectional (but actually it was bidirectional).
He was unable to postulate whether the origin of replication is unique ( know sequences from where replication is initiated every time) or occurs randomly.
DNA replication is an essential process in all living organisms for the inheritance of characters.
Read more:
Wrapping up:
Prokaryotic organisms have a single circular chromosome. Although, the replication process is a bit complicated here. Notedly, a slight variation is also reported in different prokaryotic organisms. similar to eukaryotic replication, helicase, primase, gyrase, polymerase and ligase works to unwind strands, adding primer, release tension, expanding new strand and sealing gaps, respectively.



Great work