“Regulation of gene expression is a process to control the amount of protein formed from a gene. A cell uses different mechanisms to serve this purpose. Take a look at some well-studied mechanisms here.”
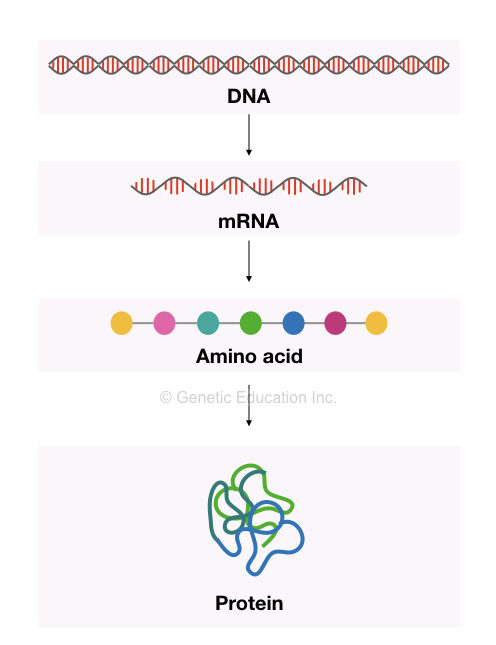
Protein synthesis is the ultimate function of a gene. A gene is a functional unit of DNA that forms various proteins by a controlled process of transcription and translation. However, at each step, a gene’s expression should be regulated.
Gene expression simply is a process of constructing an amino acid chain from a polynucleotide chain. Abruption in this process either at the transcription, translation or post-translational level results in gene dysfunction.
Put simply, it may cause the under or overproduction of a particular protein and is ideally not suitable for a cell. So controlling gene expression is pivotal for a cell. Let’s understand 12 super cool mechanisms that regulate gene expression.
Key Topics:
10 Mechanisms for Regulation of Gene expression in Eukaryotes
Transcription regulation:
Transcription is a process to construct a readable mRNA sequence. It’s the first level of control mechanism for gene expression regulation. Transcriptional machinery selectively regulates the amount of mRNA transcript synthesized.
Proteins, transcriptional factors and RNA polymerase only bind to the relaxed DNA sequences and resultantly allow transcription. This selective processing only synthesizes a required amount of transcript.
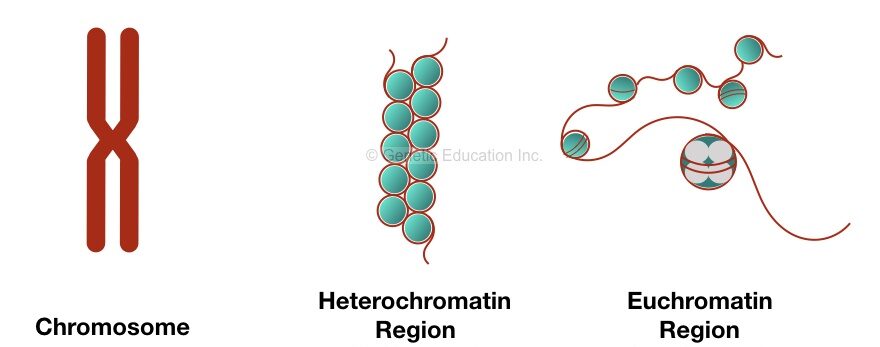
Chromatin modeling and remodeling:
Chromatin, in simple words, is a complex network of proteins (histones) and DNA having a critical role in making a gene on and off. Chromatin formation and consecutive processes in eukaryotic DNA packaging help DNA to pack on chromosomes.
Packaging is a process for tight wrapping or supercoiling of DNA which makes a gene transcriptionally inactive.
Here tightly wrapping of chromatin disallow enzymes and transcriptional factors to set on a DNA for gene expression. And consequently down-regulate a gene. Vice versa, loosely packed DNA can undergo efficient transcription.
Related article: Inside Chromatin: Definition, Structure, and Function.
The process of DNA packaging:
Not only chromatin remodeling but also other packaging processes highly regulates the entire process of gene expression. Some such steps are nucleosome assembly, 30nm fiber formation, and chromatid and chromosome arrangement.
DNA methylation:
DNA methylation is a well-documented and most prevalent epigenetic modification for gene expression. The addition of methyl groups to the DNA, specifically in the CpG region results in gene silencing.
So the amount of DNA methylation decides the amount of gene expression and final protein product. Changes in DNA methylation profile also produce serious health-related conditions and cancer.
Studies also demonstrate that methylation of DNA decides which gene to express and in which amount in different organisms.
Other chemical alterations:
Other chemical alterations like acetylation, phosphorylation, ubiquitination and hydrolysis also regulate gene expression.
Single stranded-RNA species:
Recent findings depict that several single-stranded, short, non-coding RNA species like the miRNA or siRNA also regulate gene expression in the process called RNA processing.
These RNA species process the mRNA product and perform mRNA silencing by degradation. The processing is performed by the RISC-mediated degradation pathway in the cytoplasm. You can read this article to learn more: Regulation of gene expression.
Regulatory machinery:
The transcription regulatory machinery majorly including the promotes, enhances, silences, TATA box and enzyme binding sites also play a crucial role in the regulation of gene expression. Any abruption in the activity of regulatory elements causes mRNA downregulation.
Regulatory machinery is very crucial for the initiation of transcription.
Capping and adenylation:
3’ polyadenylation and 5’ capping are crucial mRNA modifications that must be required for mRNA stability. Uncapped or non-adenylated mRNAs are processed for degradation by a cell.
However, stabilization of mRNA (with capping and adenylation) is a positive signal for initiation of translation. Thus these processes are also very important in gene expression regulation.
mRNA migration:
Protein synthesis occurs in the cell cytoplasm whilst transcription occurs in the nucleus. Henceforth, the mRNA must be transported to the cytoplasm after the completion of transcription and post-transcriptional modification.
Several proteins help the mRNA to migrate, Such proteins check the stability of mRNA to select for migration. If so, the mRNA will travel to the cytoplasm otherwise the cell degrades it.
Ribosomal recognition and translation:
Once the mRNA reaches the ribosome, the translational machinery there checks if it can be processed further or not. Usually, transcription starts with the recognition of the initiation codon AUG.
Ribosome checks the availability of AUG for synthesis. By doing so, the gene expression is regulated as well.
Post-translational modification:
Post-translational chemical modifications on the newly formed amino acid chain have been empirical too. The correct code of PTMs forms an active protein for a cell, otherwise, the partial or abrupt protein is degraded and discarded.
Read our article on this topic to learn more: Post-translational modification.
Transporting and activating a protein:
In the final step, a type of protein is packed from the amino acid chain, however, until it reaches its target action site, it’s useless.
For example, if the translational product is a cell receptor, after synthesis, it should reach a cell membrane to work as a receptor. A cell’s transportation machinery will check the stability and structure of a protein product and decide whether to send it or not.
Transporting and activating a protein is also a regulatory mechanism for gene expression. Finally, a correct protein reaches its destination and starts working, and we can say, “a gene is expressed.”
Wrapping up:
So these are some crucial, important, well-documented and studied mechanisms for regulation of gene expression. Although, it’s also notable to know that other minor processing models, which are yet not understood thoroughly, are also involved here.
Besides, extrinsic factors like the environment, food habits, addictions and climate change are factors that dysregulate the gene expression process. A gene’s ultimate goal is only achieved when a correct protein is formed and reached its site of action.
However, let me tell you that thousands of errors occur during the process but the regulatory mechanism doesn’t allow any faulty products in the process further. There are well-structured degradation mechanics as well. Dysregulation of gene expression causes serious conditions like cancer.
