“Common eukaryotic transposons are P element and retrotransposons in drosophila, Ac/Ds and Spm/dSpm in Maize, Ty element in yeast, and retrotransposons (LINEs & SINEs) in humans.”
Transposons, often known as mobile genetic elements or transposable elements can transpose or jump in the genome. Meaning, they can insert within any genetic location.
Barbara McClintock discovered transposons from Maize. Both prokaryotes and eukaryotes carry transposons. However, bacterial transposons are active but are nearly inactive in humans.
The structure of transposons varies among organisms, and thus, from organism to organism it may vary. Drosophila, yeast and maize carry active transposable elements.
In this article, I will explain to you a few well-studied eukaryotic transposons including human transposons.
Disclaimer: The content presented herein has been compiled from reputable, peer-reviewed sources and is presented in an easy-to-understand manner for better comprehension. A comprehensive list of sources is provided after the article for reference.
Stay tuned.
Key Topics:
Transposons in Drosophila
15% of the drosophila genome comprises mobile genetic elements. Two common types are P elements and retrotransposons. These play a crucial role in their genome evolution, creating mutations and gene regulation.
P elements
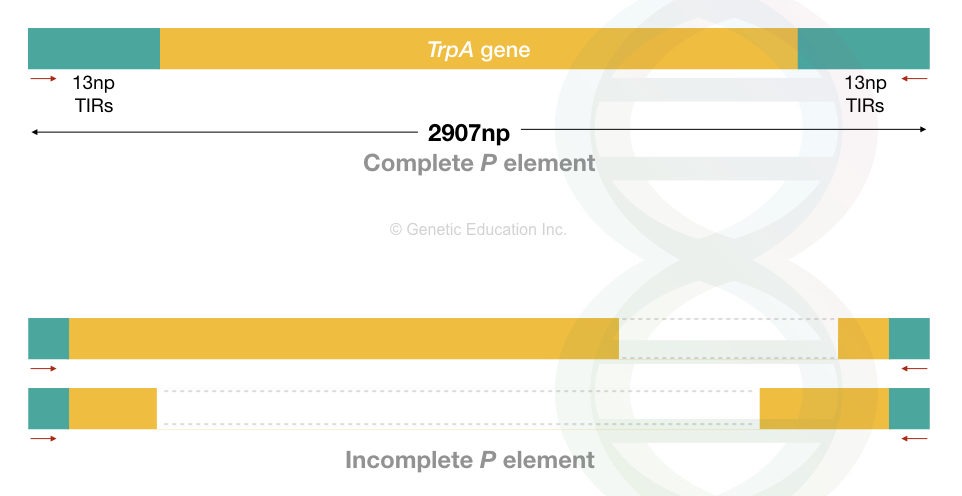
The drosophila P elements are 2907 bp in length. However, the size may vary. These are autonomous DNA transposons and follow a cut-and-paste transposition mechanism.
That means it excises from one location, creates a gap and inserts to another location. Like the other transposons, it contains a gene body, inverted terminal repeats (31 bp) and target site duplication (8 bp).
Furthermore, P elements in the drosophila are either complete P elements with the transposase gene or incomplete P elements, without the transposase gene.
The complete P elements can encode a transposase gene, which means it can perform transposition efficiently. However, due to the lack of transposase gene and terminal repeats, incomplete P elements can not effectively perform transposition.
Because of the incomplete nature of P elements, it may vary among drosophila. The incomplete transposons are non-autonomous.
The class II transposons— P elements (complete) contain 4 exons and 3 introns and produce transposase and repressor by splicing and alternative splicing, respectively.
Complete P elements insert themself into functional genes and produce either mutations or altered gene expression.
The P element transposition can be understood using the P-M hybrid dysgenesis mechanism.
P-M hybrid dysgenesis
In this process, flies from two different genetic backgrounds P cytotype and M cytotype are crossed to investigate the transposon-mediated transposition. This helps scientists to determine how the P element moves within the genome and how genetic diversity is produced.
Flies comprising the P elements with a regulatory mechanism are known as a P cytotype. Meaning, that the P-element-mediated transposition is highly regulated by suppression.
Now this P cytotype is maternally inherited and offspring with the present cytotype are protected from harmful transposition activities.
On the contrary, the M cytotype flies do not carry the P element but are susceptible to P element transposition.
Look at the image below to understand the mechanism.
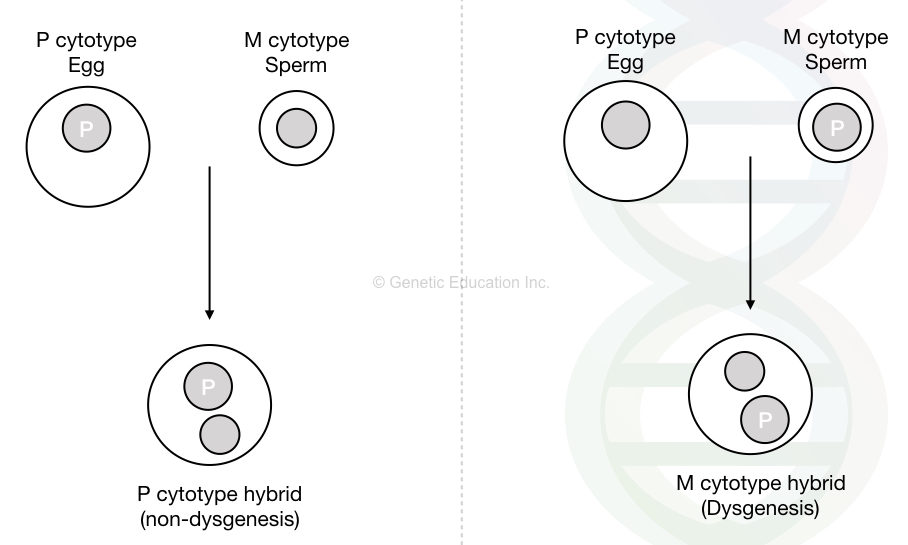
Now, here the hybrid dysgenetic does not occur if the P cytotype female (carrying the P element) is crossed with the male of M cytotype male. On the contrary, hybrid dysgenesis occurs if the P cytotype male is crossed with the M cytotype female.
Now in the second case with dysgenesis, the introduction of the P element by a male is not suppressed as it lacks a maternally inherited P cytotype. These offsprings show the transposition of P elements without any regulation.
The present experiment also showed that the P element also encodes a suppressor protein, for regulating the transposition but it accumulates in the cytoplasm and is inherited maternally.
This theory is known as P-M hybrid dysgenesis.
Interestingly, studies showed that the P elements were not present in drosophila before the 1950s. That means P elements are evolved through the process of natural selection.
The reason is still unclear to scientists, however, one hypothesis suggests that P elements were introduced in drosophila by the viruses that naturally infect them.
In addition, not only D. melanogaster but other distantly related species also carry P elements in their genome. P elements are further a good tool for genetic engineering and research. Scientists employ it to understand genetic instability and its evolutionary impact.
Furthermore, it is applied for genetic modifications, mutagenesis and gene editing experiments.
Drosophila retrotransposons
Major transposon portions in drosophila carry retrovirus-like elements, known as retrotransposons. The class I retrotransposons are structurally similar to the yeast Ty elements and retrovirus.
It ranges between 5Kb to 15Kb. It has a long terminal repeat oriented in the same direction. And much like the other transposons, it also carries the target site duplication on both ends.
In drosophila, the present class of transposons is divided into two types– copia and gypsy. The difference is due to the target site duplication. The copia contains 5 nucleotides of TSD whereas gypsy contains 4 nucleotides of TSD.
Notedly, the target site duplication has a similar orientation. It is also evident that copia and gypsy elements also carry a gene for reverse transcription with the transposase.
The reverse transcriptase enzyme governs the conversion process from RNA to DNA. In the natural system, the mRNA is formed from the DNA, and here, after that, the DNA is synthesized again from the mRNA through reverse transcription.

A short note for copia and gypsy is given below.
Copia:
- LTR (Long Terminal Repeat) retrotransposon.
- Contains genes for reverse transcriptase and integrase.
- Follow the copy-and-paste mechanism (replicative transposons).
- Reverse transcribed into DNA and inserted into a new genomic location.
- Disrupt gene function and regulation.
Gypsy:
- LTR- retrotransposon.
- More like the retrovirus.
- Act like a virus and move between cells as well.
- Carry genes for reverse transcriptase and envelope (env gene provides the capacity for extracellular transmission).
- Disrupts gene function and regulation.
Transposons in maize:
Maize is a classic model to study transposable elements as 80% of its genome is made up of only transposons. Transposons were first discovered in maize by Barbara McClintock.
Two common types of maize transposons are Ac/Ds and Spm/dSpm (or Spn/En)
Ac/Ds elements
Ac elements are activators, present throughout the maize genome and are autonomous. It can independently jump within the genome. They are structurally similar but not identical.
It is 4563 bp in size with 8 nucleotides long direct terminal repeats and 11 nucleotides long inverted terminal repeats. It possesses sequence homogeneity and is interspersed within the genome.
Ac elements can not replicate on their own. What it does is! It is copied by a cell’s natural replication process. It is excised from one location, replicates with the cell’s DNA and generates another copy.
However, a mutation within the transposase gene of the Ac element can make it inactive.

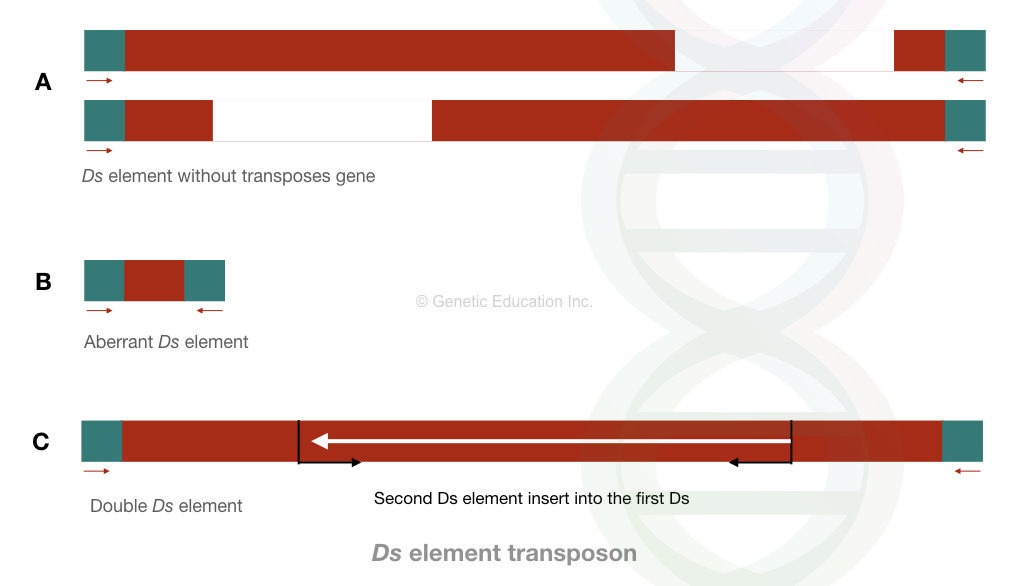
Another element in this class is the Ds element. Ds is a “dissociate element” and is structurally and functionally different from the Ac elements. It’s non-autonomous, which means it can not move on its own, it needs activators.
It is an aberrant kind of element that lacks the main gene body or sequence. For instance, some Ds elements contain terminal repeats from the Ac elements, they lack their own.
In the double Ds DNA case, one Ds element is inserted into another one. These are responsible for chromosomal breaks. Interestingly, Barbara McClintock identified these elements and was awarded the Nobel Prize.
The Ac/Ds system transposon sequences were sequenced by Federoff et al. (1983) through Wx1 gene insertion. Notedly, these elements are also present in Rice, Arabidopsis and Tobacco.
Spm/dSpm
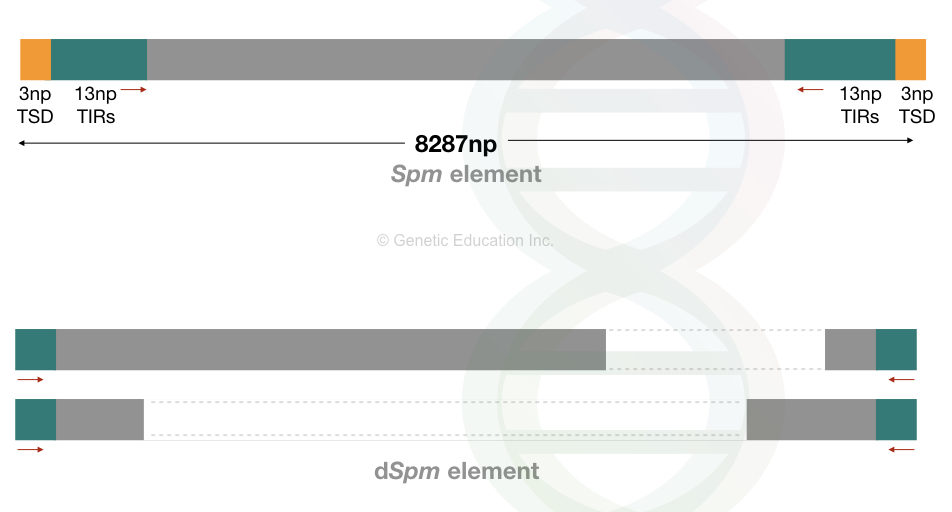
Spm means suppressor mutator, a type of autonomous transposon in the maize genome. While the dSpm is deleted or defective suppressor mutator, a type of non-autonomous maize transposon.
Spm is 8287 bp long TE with 13 nucleotides long terminal repeats and 3 nucleotides long target site duplication.
Conversely, the dSpm elements can not move on their own. They lack a functional transposase gene due to the deletion within the transposase sequence. And thus, are smaller in size than Spm.
The Spm controls the activity of dSpm upon interaction. Ac and Spm elements are epigenetically regulated.
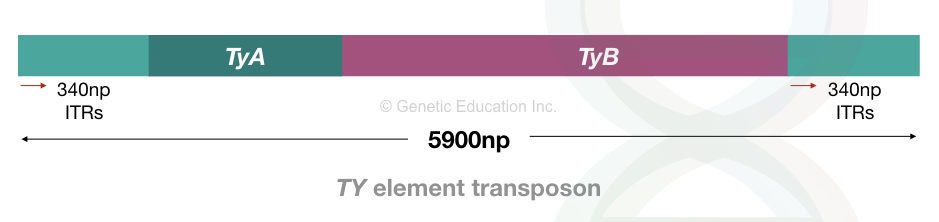
Ty elements in Yeast
Ty transposons are retrotransposons, like tranposons in other eukaryotes. It is present in Baker’s yeast (Saccharomyces cerevisiae) and is one of the well-studied eukaryotic transposons.
It is functionally similar to the retrovirus as it is reverse-transcribed into DNA through an RNA intermediate. The Ty element is 5900 bp in size and consists of 340 bp long terminal repeats.
These are the inverted terminal repeats and located on both 3’ and 5’ of the TE and are known as δ (delta) sequences.
Studies indicate that the Ty elements most frequently jump in the AT-rich genomic region due to the presence of an AT-rich sequence as target site duplication. The TSD is 5 nucleotides long.
With 35 copies within a single genome, the Ty elements contain TyA and TyB genes encoding structural and catalytic proteins, respectively. The TyA gene is similar to the retroviral ‘gag’ gene and encodes structural particles.
The TyB gene is similar to the ‘pol’ retroviral gene that encodes for reverse transcriptase and protease and integrase. Ty elements are replicative transposons and follow a “copy-and-paste” mechanism.
As aforementioned, using the RNA intermediate; it synthesizes the DNA and inserts it into a new location through the reverse transcription process.
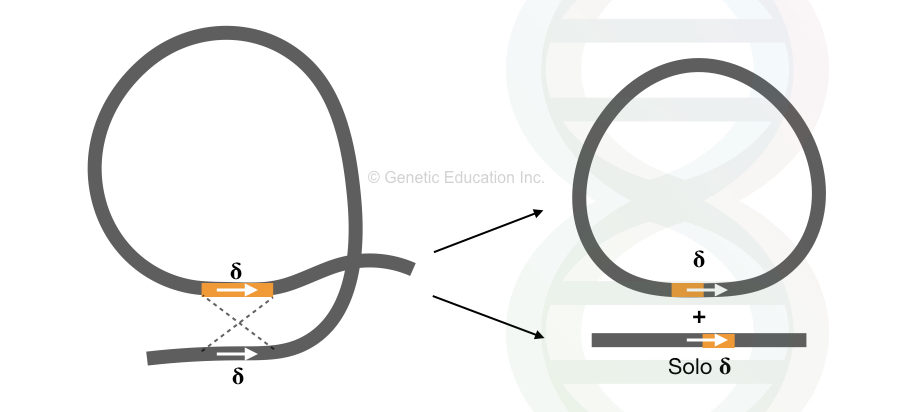
As shown in the image, the recombination between two Ty elements generates solo δ (delta) sequences. However, one circular byproduct of unknown significance is also generated.
Ty elements are divided into subfamilies Ty1, Ty2, Ty3, Ty4 and Ty5. Check out the table for more specifications.
| Ty element subfamily | Specification |
| Ty1 and Ty2 | Well-studied, most common, functionally similar and closely related. |
| Ty3 | Closely related to retrovirus with a slightly different gene structure. |
| Ty4 | Not abundant but structurally similar to Ty1 and Ty2. |
| Ty5 | Inserts specifically near telomere and non-coding regions. |
Much like other transposons, Ty elements can be inserted within a functional gene and make it inactive. Due to the resemblance of the Ty element, its TyA and TyB gene with the retrovirus, it is believed that they evolved from the retroviruses.
Human transposons
Approx. 45% of the human genome is made up of transposons or transposon-derived sequences. However, they are no longer active. A major portion of the human transposons is retrotransposons.
Only 3% of TEs consist of DNA transposons as Tc1/mariner family TEs.
The retrotransposons follow the copy-and-paste transposition mechanism (replicative transposition). It prepares the DNA through RNA transcript and inserts it into a new location.
Two common types of human retrotransposons are LINEs (Long Interspersed Nuclear Elements) and SINEs (Short Interspersed Nuclear Elements). We already discussed these two in our introduction article.
You can read it here: What are Transposons?- Introduction, History and Types.
I’m just giving you a brief overview here.
| LINEs | SINEs |
| Long Interspersed Nuclear Elements | Short Interspersed Nuclear Elements |
| ~6000 bp in length | 100 to 300 bp in length |
| Autonomous | Non-autonomous |
| Replicative transposons | Replicative transposons but depends on LINEs |
| Encodes reverse transcriptase and endonuclease | Do not encode proteins |
| Cover 17% of the genomic content | Cover 10% of the genomic content |
| Example is L1 | Examples are MIR and Alu. |
Both LINEs and SINEs are the type of non-LTR retrotransposons. Meaning, they lack long terminal repeat sequences. Notedly, their transposition creates new mutations, affects gene function and expression and drives evolution.
Wrapping up:
Transposable elements are significant forces in evolution that help produce new alterations. However, they can also produce some harmful traits as well. Scientists are using it as a genetic research tool, particularly in plant research.
TEs are used in artificial mutagenesis and help in genetic engineering and gene therapy experiments. Fortunately, they are genetically and epigenetically silenced in humans.
I hope you like this article. Share it and subscribe to Genetic Education.
Sources:
Wells JN, Feschotte C. A Field Guide to Eukaryotic Transposable Elements. Annu Rev Genet. 2020 Nov 23;54:539-561. doi: 10.1146/annurev-genet-040620-022145. Epub 2020 Sep 21. PMID: 32955944; PMCID: PMC8293684.
Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187-215. doi: 10.1146/annurev-genom-082509-141802. PMID: 21801021; PMCID: PMC4124830.


