“FISH- Fluorescence In Situ Hybridization allows the direct study of a sequence located on the chromosome. It has been used for insertion and selection studies. Let’s see what FISH is and how it works.”
Broadly, cytogenetics and molecular analyses are two categories of genetic techniques that can study chromosomes and associated aberrations, and DNA or gene sequence, respectively. Conventional karyotyping, and PCR and sequencing are examples of each technique.
Each technique has its own importance. Conventional cytogenetic investigations like routine karyotyping encounter abnormalities at a chromosomal level. Such a process often needs subsidiary techniques like chromosome banding. For example, GTG banding for numerical analysis and NOR banding for trisomy 21.
However, the conventional cytogenetic approach is not sufficient enough to study smaller deletions and duplications due to low resolution. On the side, molecular techniques enable us to study only smaller sequence variations like SNP, a few nucleotide copy number variations, etc.
But intermediate-level indels like a few thousand basepair deletions or insertions can’t be studied by either method. To overcome this problem molecular cytogenetic techniques like FISH are developed.
FISH can determine sequence variations directly from the chromosome and provides information regarding the location of the alterations. In this article, we will discuss only FISH, what is it and how it works, along with the process, protocol, advantages and applications.
Stay tuned.
Read our previous article on cytogenetics: A Brief Introduction To Cytogenetics [Karyotyping, FISH and Microarray].
Key Topics:
The overall idea of FISH:
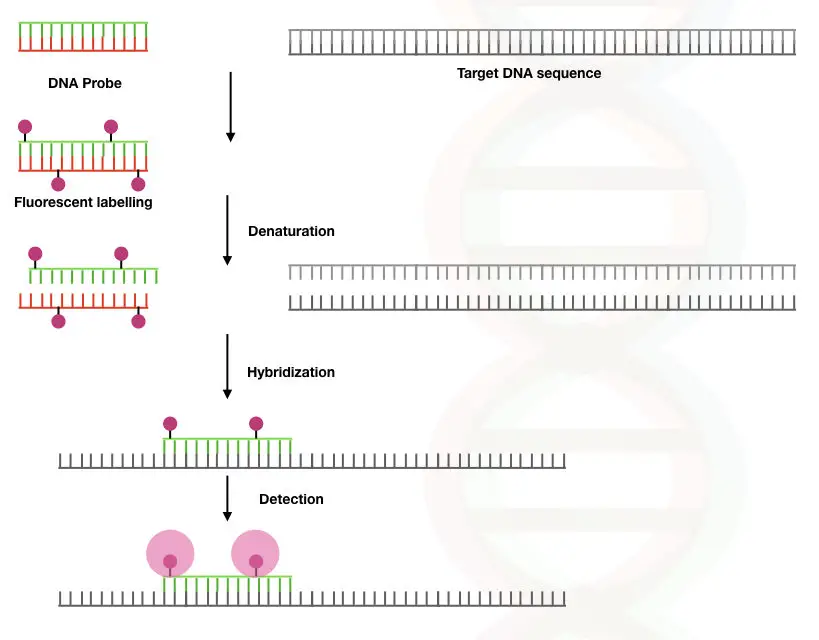
The FISH method is based on the phenomenon of the denaturation and renaturation of DNA duplex. The DNA is a stable duplex, under normal conditions hydrogen bonding between two strands (two between adenine and thymine; and three between cytosine and guanine) makes it stable.
Although the duplex can be denatured using physical agents such as heat or chemicals when conditions are favorable, DNA becomes renatured.
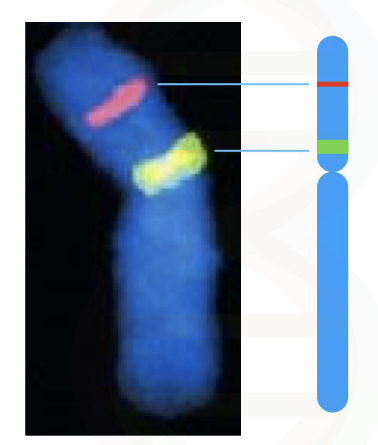
When DNA on the chromosome is denatured, the fluorescently labeled complementary DNA probes hybridize with it and emit fluorescence, consequently. The hybridization is visualized under the fluorescent microscope.
The technique is an advanced version of the cytogenetic analysis and employed for gene mapping, identification of major deletion or copy number variations, disease diagnosis, medicines, and species identification.
What is Fluorescent in situ hybridization?
In a FISH, larger deletion/insertion can be mapped by hybridizing fluorescent probes complementary to the sequence on the chromosome, directly from a cell.
The conventional karyotyping approach, as aforementioned, is a tedious, time-consuming, error-prone and costly method, only finds numerical abnormalities and needs utmost expertise in the field. Such investigations are not sufficient for copy number variation studies.
FISH is a ‘molecular cytogenetic technique‘ in which using molecular probes, any type of chromosomal abnormalities can be encountered precisely by hybridization. The karyotyping takes at least 3 to 4 days to complete the entire process while the FISH method is rapid, one can get results within a day.
The interpretation part required one-fold lesser expertise.
- Fluorescence: light emitted by a molecule
- In situ: on the original place or on the appropriate location (in our case, in a cell on the glass slide).
- Hybridization: here, binding of two complementary strands.
Principle of FISH:
Directly in a cell, by chemical treatment and using a higher degree of sequence complementation, probes of DNA or RNA are hybridized on a chromosome(s). The fluorescent-labeled probes when finds their complementary sequence (which is a sequence of our interest), hybridize and emit fluorescence. Signals are recorded and investigated using a fluorescent microscope.
The unbound probes are washed off to avoid unwanted signals from the site of hybridization. The technique was developed in the year 1980. The whole principle is graphically explained here,
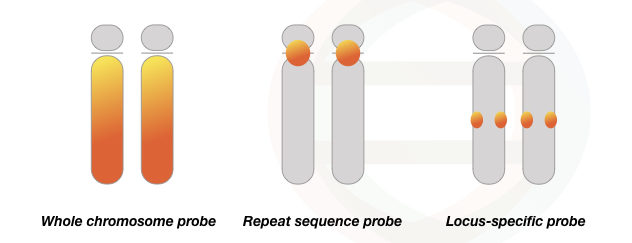
Types of probes used in the FISH:
FISH is a diverse technique, that needs separate probe preparation to inspect each sequence. Thus assay and probes vary from experiment to experiment, however, the basic process and principle remain the same. Four well-established probe types are centromere probes, locus-specific probes, whole chromosome probes and telomere probes.
Whole chromosome probe:
The whole chromosome probes apply in multi-color FISH and spectral karyotyping. It is a mixture of probes that binds to the entire chromosome length and thus different chromosomes are colored or labeled with different colored probes.
Each probe is derived from a single type of chromosome and binds to that particular chromosome only. It is employed in chromosomal rearrangement studies.
Locus-specific probe:
The locus-specific probes are used in the study of a particular gene or DNA sequence of interest. Consequently, it allows the identification of the gene location or target chromosomal region. It can effectively find chromosomal anomalies like deletion or duplication of a gene.
In addition, specially designed multi-locus specific probes enable the detection of multiple DNA/gene sequences on different chromosomes.
Alphoid or centromeric probes:
Alphoid probes use the repetitive nature of chromosome sequences. The centromeric and telomeric regions of a chromosome comprise highly repetitive sequences. Such sequences are used to develop Alphid probes which are often known as repeat sequence or alphoid-centromeric probes.
These types of probes are used in numerical chromosomal anomalies study and find monosomy, disomy or trisomy.
Take a look at the figure to know how different probes work.
Examples of some commercially available probes for different FISH assays are given below,
| Probe name | Used for | Location |
| HER-2/CEP17 | Breast cancer | 17q11.2-q12 |
| TOP2A | Breast cancer | 17q21-22 |
| ERG | Prostate cancer | 21q22.2 |
Note:
Different probes are nowadays available for different chromosomal anomalies, probe designing is not required for performing any FISH experiment.
Process of FISH:
FISH is indeed a simple and robust hybridization technique that requires only two things- target sequence and specific probe. Steps are elaboratively explained here.
Selection of target sequence
Target selection is a critical step. Keep in mind that FISH can effectively locate chromosomal regions of more than several thousand basepairs. So select a target having this property. However, the region of interest might be any deletion, duplication, translocation, disease-related gene or a DNA sequence.
The target is selected using computational analysis.
Design the probe
Once the target sequence is selected, based on the data of the sequence we wish to study, the probe is designed. The probe is a single-stranded sequence of DNA or RNA complementary to our gene of interest.
The polynucleotide chain which we are utilizing as a probe is then labeled with the fluorophores. Generally, the direct labeling method is used for probe generation, in which the fluorophores are directly attached to the nucleotides.
The tagging of fluorophores on the nucleotide sequence can be done using either the nick translation method or PCR. Read our article on preparing probes: DNA Probes: Labelling, Types And Uses.
Sample preparation
Both fixed or unfixed samples can be used in FISH. Paraffin-embedded tissues, FFPE tissues, tumor cells, cell culture, or chromosomal suspension are some common sample types for FISH analysis.
In the very first step, by chemical treatment, cells are permeabilized for probes to enter, afterward, under favorable conditions, probes are allowed to hybridize.
In the next step, the cells are permeabilized using the chemical treatment for allowing the probe to enter into cells and hybridize on chromosomes. However, for some specialized assays like fiber-FISH, the chromatin fibers are extracted and stretched on a slide using the fluid-flow method. Fibre-FISH has greater resolution.
Denaturation:
In the next step, the sample is denatured.
The DNA is a double helical structure and more stable. For making hybridization possible, we need to denature the sample so that our probe can bind to the DNA sequence. A higher temperature or denaturing agents are used for generating single-stranded target DNA for hybridization.
The DNA is denatured using heat or alkaline agents.
Hybridization process
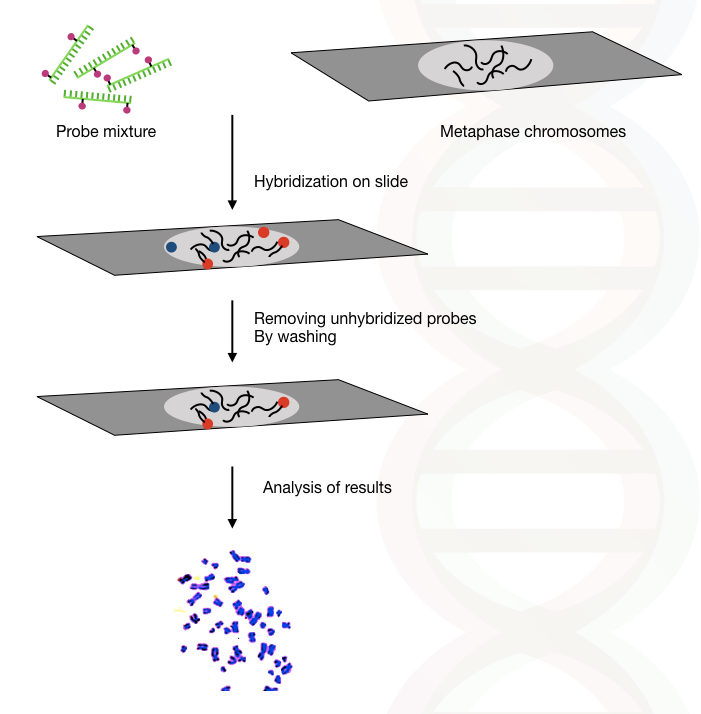
Interphase or metaphase chromosomes are the best choice for performing FISH efficiently. After the sample is prepared, the probe mixture is applied on the surface of the glass slide having the sample. Along with it, short DNA fragments are added to block the repetitive DNA hybridization with the probe.
Once hybridization completes, we need to wash the slide in order to wash off partially and incompletely hybridized probes. A ready-to-use wash buffer is recommended to wash the slide. Wash the slide as instructed by the manufacture.
In the next step, the slide is incubated for 12 hours for hybridization to occur. After that, the results are analyzed under the fluorescent microscope.
Results and interpretation:
Scientists visualize the slide under the fluorescent microscope, if the probe hybridized properly on its complementary sequence, it emits two fluorescent signals. The signals are clearly observed, however, expertise is required to interpret the results.
A brief overview of the whole process is explained hypothetically here in the figure.
Related article: Genetics Basics: A Beginners Guide To Learn Genetics.
A general protocol for FISH:
Materials and instruments:
Fluorescent dye or fluorescent-labeled probe complementary to our sequence of interest, sample specimen, fluorescent microscope, alkaline agent, SSC buffer, 10mM HCl, hybridization solution, ethanol, coverslip, slide, heating block, humid chamber and incubator.
Process:
- Prepare the slide with the cell suspension.
- Incubate the slide with 200µl RNAse solution for 1 hr at room temperature.
- Wash the slide with 2X SSC for 4 to 5 minutes, followed by 10mM HCL rinsing.
- Add 200µl pepsin and incubate for 8 to 10 minutes at room temperature.
- Rinse slide with distilled water and then with 2x SSC.
- Place the slide in the paraformaldehyde solution for 5 to 10 minutes for fixing and wash it with the same SSC buffer.
- Now perform dehydration to dry the slide with 70%, 80%, and 90% of ethanol each for 2 minutes.
- Place the slide for some time to air dry. Our slide is ready for hybridization.
- Add 30µl of hybridization solution on a slide, heat it at 65 to 70°C for 10 minutes and cool it by placing it on ice.
- Cover the slide with a coverslip and again heat it 65 to 70°C for 5 minutes for denaturation.
- Place the slide at room temperature for the hybridization of probe and DNA.
- Place it in a humidity chamber overnight at 37°C.
- Next morning, wash the slide with SSC buffer (2X) and then wash with 0.1X SSC buffer at 40°C and remove the coverslip.
- Give a final wash to slide with 2X SSC buffer at 40°C and block it in a blocking buffer for 30 minutes.
- Add antibody to the slide and incubate it for an hour and with it with 2X SSC buffer.
- If needed repeat the washing step followed by DAPI counterstain.
- Air-dry the slide and visualize under the fluorescent microscope.
Note:
The protocol may vary from lab to lab, minor modifications are required to achieve good FISH results. The protocol is originally adopted from Sigma-Aldrich.
Different types of FISH:
Depending upon the requirement of the study, different variations of the native FISH are available nowadays.
M-FISH:
The M-FISH known as multicolor FISH uses different colored probes for different chromosomes. Broadly, it is used in the characterization of different chromosomes and numerical chromosomal abnormalities.
Q-FISH:
Known as quantitative FISH is used for the quantification of the genetic material hybridized by the probe. The fluorescent intensity is measured for quantification thus it is used in the study of telomere and aging, cancer and gene expression.
Flow-FISH:
Flow cytometry FISH is a variation of FISH used for the quantification purpose which measures fluorescent emitted from every single cell detected and measured.
e-FISH:
A computation tool used for the prediction of the outcome of the FISH experiment is called electronic FISH. It is a BLAST-based program that utilizes the sequence information for in silico estimation of the hybridization process.
Fiber-FISH:
Using chromatin fiber or DNA fiber, the high-resolution gene mapping can be done using the Fiber-FISH method. The present modification even permits the identification of the DNA fiber less than 1000bp.
Thus it is widely used in the gaps and overlap fragment analysis, assessment of duplication and other copy number variation detection which can not be detected by the conventional FISH method.
Using the salts or solvents, the chromatin fibers are released and fixed on the glass slide, instead of the whole chromosomes. The probe hybridization is done directly on the glass slide containing the chromatin fibers.
ACM-FISH:
The ACM-FISH is a variation of the multicolor FISH especially designed for sperm cells.
The ACM stands for alpha (centromere), classical and midi satellites of chromosome 1 for detection of duplication and deletion on the chromosome. It is used for the detection of the role of chromosomal damage in the development of infertility in males.
catFISH:
The method is a type of RNA-FISH used to study the neurons associated with abnormal cognitive behavior. Cellular compartment analysis of temporal(cat) is abbreviated as catFISH.
MA-FISH:
Microfluidics-associated FISH uses microfluidics for improving the performance of the FISH. It increases the hybridization efficiency as well as decreases the time consumption thus it is used in bread cancer for detection of the HER2 gene mutation.
COMBO-FISH:
COMBO-FISH stands for combinatorial oligonucleotide FISH used for the detection of homopurine or homopyrimidine region of the genome. The homopurine and homopyrimidines cover approximately 2% of the human genome.
The probes are designed to hybridize on these regions of the genome and create a triple helical structure with it by binding with the DNA duplex.
This unusual triplet structure is located using the fluorescent signals having the homopurines or homopyrimidines, this information is used for the 3-D (three-dimensional) study of the human genome.
One of the major advantages of COMBO-FISH is that we do not need to denature the sample prior to hybridization, thus, reduce the complexity of the FISH assay.
SM-RNA-FISH:
Single-molecule RNA-FISH is employed for the quantification of gene expression from the tissue sample, using the hybridization method. Here, instead of DNA, RNA is used as a target for probe hybridization.
Note: a special type of FISH is applied for the temporary gene expression pattern within cells by using the RNA as the template for the FISH.
Advantages of FISH:
Conventional karyotyping relies on cell culture which is tedious, sensitive to contamination and a time-consuming process. The major advantage of FISH is that it doesn’t require cell culture. This makes the technique more powerful, rapid and accurate.
Conventional karyotyping techniques require dividing cells to arrest at metaphase but the FISH can effectively work with non-dividing and diving cells too. Therefore, chromosomes from solid tumor cells can be studied.
Karyotyping techniques are restricted as it depends on chromosome banding. But in FISH, using multiple probes, multiple hybridization sites have been analyzed using different fluorophores. In fact, FISH works on not only metaphase but also on interphase chromosomes too.
The cross-reactivity rate of the FISH probes is also very low.
It has a higher signal-to-noise ratio.
Paraffin-embedded, frozen tissue and cultured cells are used for FISH analysis, besides this, both types of cells (non-dividing as well as dividing cells) are applicable in FISH.
Applications of FISH:
Preliminary investigations such as specific chromosomal abnormality, and numerical or structural alterations like deletion, duplication or translocation can be studied effectively through FISH.
Varients like SKY or M-FISH can find non-random and new genetic abnormalities associated with chromosomes.
Spectral karyotyping (SKY) or multicolor FISH (m-FISH) is a modification of native FISH in which using the different colored sequence-specific probe each chromosome can be painted and chromosomal rearrangements can be encountered. However, smaller deletions and/or duplications can not be identified using Spectral karyotyping because of the restricted resolution.
It is used in gene and genetic mapping. We have covered an article on gene mapping. Read it here: A Brief Introduction to “Gene Mapping”.
FISH studies, identifies and characterizes a marker chromosome and is often employed to determine a specific breakpoint from where translocation occurred.
One of the most fascinating applications of quantitative FISH is in the monitoring of disease progression.
If the conventional karyotyping fails to find out any abnormalities, using the interphase FISH (which provides higher resolution), those types of abnormalities can be identified.
Moreover, the FISH is used in the study of three-dimensional chromosomal structure and organization.
It is practiced for monitoring the success of transplantation, for example, bone marrow transplantation.
Conclusion:
The fluorescence in situ hybridization technique is capable of detecting larger copy number variation efficiently. Scientists are now applying different variations of FISH for different cytogenetic applications. One of them is comparative genomic hybridization. CGH is used for quantitative detection of copy number variations.
Although FISH is a highly versatile and rapid method, one should learn the basic chromosome preparation and karyotyping methods to boost their knowledge and expertise.
I strongly recommended learning basic karyotyping. Read our series of articles on cytogenetics:
- A Karyotyping Protocol For Peripheral Blood Lymphocyte Culture.
- What Is The Role Of RPMI 1640 In Karyotyping?
- Role Of L-Glutamine In Karyotyping.
Sources:
- Emanuela V & Joanna M. “FISH glossary: an overview of the fluorescence in situ hybridization technique.” Biotechniques. 2018; 45: 385-409.
- Ratan ZA, Zaman SB, Mehta V, Haidere MF, Runa NJ, Akter N. Application of Fluorescence In Situ Hybridization (FISH) Technique for the Detection of Genetic Aberration in Medical Science. Cureus. 2017;9(6):e1325. Published 2017 Jun 9.
- O’Connor, C. (2008) Fluorescence in situ hybridization (FISH). Nature Education 1(1):171 (link).
Thank you this was very helpful 🙂