“Single Nucleotide Polymorphism is a single nucleotide alteration within a DNA sequence that produces different alleles. Explore the concept of SNPs in this article.”
The human genome is mysterious. With approximately 3.2 billion bases, the human genome is an ocean of information. Unfortunately, around 23,000 genes are yet known to us.
This comprises around 3% part of the genome and the rest is still a question to us. Replication helps copy the entire set of DNA present in the cell. The process is highly efficient and errorless (mostly!).
A mutation occurs when this efficient machinery messes up things. Usually, the cell takes responsibility and repairs it. Oftentimes, it remains unrepaired and causes problems.
An SNP is one such mutation that frequently occurs within our genome thousands to millions of times. SNPs are now a hot topic in genetic research and everyone is talking about it because of their tremendous advantages.
In this article, I will explain SNPs, their basics, types, nomenclature techniques and databases. The present article will help you learn the concept of ‘mutation’ and its association with diseases more accurately.
Related article: Genetic Mutations- Definition, Types, Causes and Examples.
Stay tuned.
Key Topics:
What is Single Nucleotide Polymorphism?
Single Nucleotide Polymorphism (in short) is known as SNP. SNP is the widely accepted and known term to introduce single nucleotide polymorphism. It is pronounced as ‘snip’ or ‘snips.’
SNPs are crucial genomic alterations that gained much attention after 1980. However, now in 2023, it’s a major focus of genetic research. Various SNPs are involved in not only disease development directly but are associated with complex conditions, indirectly.
Single= One
Nucleotide= Base+ suger+ phosphate,
Polymorphism= Alteration
A nucleotide is a structure having a nitrogenous base, sugar and phosphate. One nucleotide is joined to another nucleotide with a phosphodiester bond and prepares a long polynucleotide chain of the DNA.
Nitrogenous bases A, T, G and C are the constituents of a nucleotide. Whenever any alteration occurs within the polynucleotide chain, it results in mutation. We also know it as polymorphism.
If any single nucleotide is altered, for instance— if A nucleotide is replaced by a G nucleotide, the condition is referred to as a single nucleotide polymorphism/single base change/ single nucleotide alteration, etc.
All these terminologies explain the same condition– SNP.
On average, an SNP occurs after every 1000 nucleotides. Thus approximately 4 to 5 million SNPs are present in our genome. SNPs occur during the process of replication and as the replication is a never-ending process, thousands to millions of SNPs originate every single day.
However, the majority of single-base alterations are repaired by our well-equipped DNA repair mechanisms. According to a recent report, scientists have identified approximately 600 million SNPs from various populations worldwide.
Those numbers are huge! And if it is associated with health conditions, the numbers are scary too. Right! Let me tell you that a large number of SNPs don’t have any direct role in disease development.
The reason is! Majorly, SNPs are present in the non-coding genomic region and in intervening gene regions. Thus, largely it isn’t a matter to worry about, in general. However, in some cases, SNPs in non-coding regions can cause a disease as well.
Thalassemia, Sickle cell anemia and Cystic fibrosis are a few examples in which SNPs are involved in causing a disease.
If an SNP occurs in a coding gene segment, it alters the genetic code and resultantly alters the protein encoded by that gene. So an SNP within the coding segments or a gene is a matter to worry about! Certainly.
Definition:
Single nucleotide polymorphism (SNP) is defined as any single nucleotide alteration within a DNA sequence, gene, or genome, known as SNP.
Read more: Trinucleotide Repeat Expansion Disorders 101: Mechanisms and List of Disorders.
SNP Types:
Now, before moving ahead, we need to understand different types of SNPs. Starting with the coding and non-coding SNPs.
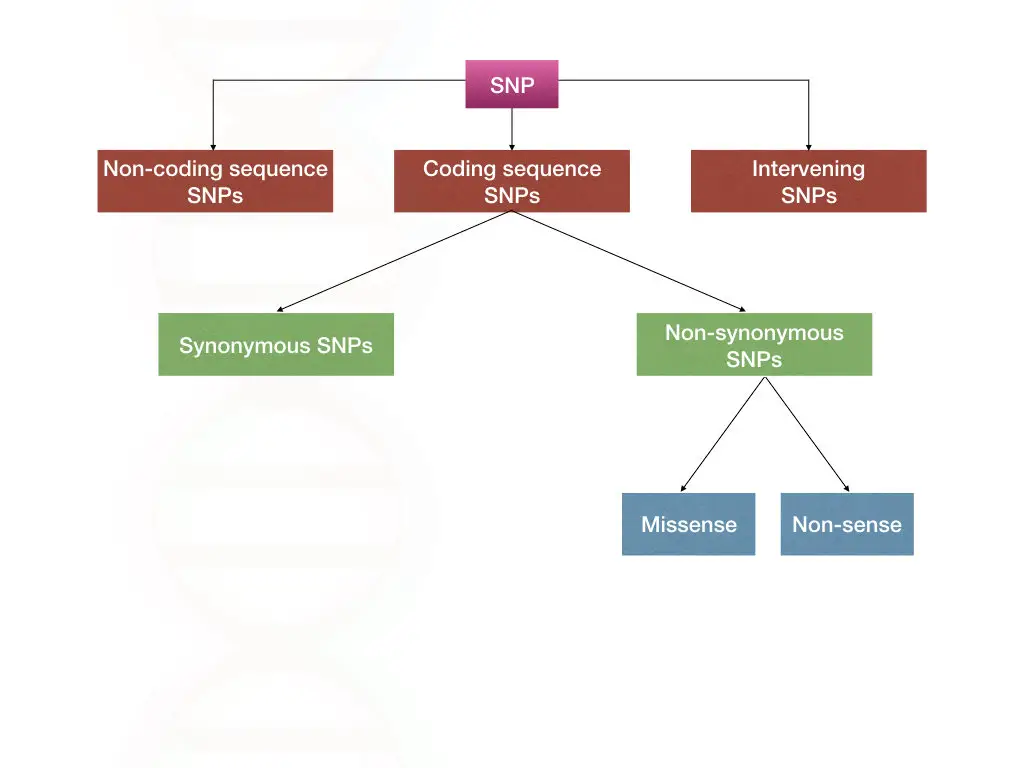
SNPs in non-coding sequence:
When a single base change occurs within the non-coding regions, is known as non-coding SNPs.
SNPs in coding sequence:
When a single base change occurs within the coding region of the gene, is known as coding SNPs. These types of SNPs are further divided into synonymous SNPs and non-synonymous SNPs.
Synonymous SNPs:
These are the types of SNPs that do not affect the amino acid sequence or final protein product.
Non-synonymous SNPs:
These are the SNPs that do change the amino acid sequence and the final protein product. It can likely affect protein function. A single nucleotide missense and nonsense mutations are categorized into non-synonymous SNPs.
Here, there might be two scenarios. The protein function can be altered or can not be altered. Based on that, the non-synonymous SNPs are further divided.
Conservative SNPs:
It is a type of non-synonymous SNP. Here amino acid change would not affect the final protein product. Meaning the amino acid change will remain similar to the original.
For example, the arginine amino acid is encoded by CGC, CGA and CGG codons. If the A nucleotide is replaced by the C or G, the arginine amino acid will still be synthesized.
Non-conserved SNPs:
Now, when non-conserved SNP occurs it will change the amino acid and consequently affect the final protein product. For example, the AUG codon encodes methionine. If SNP replaced a G nucleotide with any other nucleotide, methionine would not be produced.
So non-conserved SNPs directly affect the protein function. It results in either abrupt, abnormal or truncated protein products.
Intergenic SNPs:
Intergenic SNPs occur between the coding regions of the gene. Meaning, it can alter the regulatory elements of a gene and influence gene expression. For example, Beta thalassemia IVS 1-1 mutation is present in the intervening sequence of the beta-globin gene.
Transition SNPs:
When SNP replaces a purine base with another purine base or pyrimidine with another pyrimidine is known as transition SNP. For example A to G and C to T and vice versa.
Transversion SNPs:
When SNP replaces a purine base with pyrimidine and vice versa. For example A to T or C to G etc.
Splice site SNPs:
These SNPs are present in a gene’s intron and exon junction and potentially affect the splicing process.
Insertional SNPs:
In an insertional SNP, a nucleotide is inserted into a DNA sequence. Note that here, a fresh/new nucleotide is added, it doesn’t replace any nucleotide.
Deletion SNPs:
In deletion SNPs, a nucleotide is deleted from a gene or a sequence. Check out this image for a better understanding.
Pathogenic SNPs:
Pathogenic SNPs increase the risk of developing a particular disorder. These SNPs can cause a genetic disease, abnormal health condition or complex abnormalities.
Non-pathogenic SNPs:
Non-pathogenic SNPs don’t have any impact on a person’s health and do not cause any disease.
SNP nomenclature:
Whenever you read some article you may come across terminologies like rs321012 or c.122A>T. These are the two styles of writing an SNP. We can say, it’s a nomenclature technique.
At Genetic Education Inc. we present literature in a more understandable manner. How can we avoid explaining this? Let’s understand the SNP nomenclature.
Firstly, whenever you come across such terms as c.122A>T. Or rs321012, make it clear that the author is talking about some SNPs. Here,
C. is the codon. This signifies the coding region.
122 is the location on a gene.
A>T is an alteration. Here A nucleotide from the original sequence is replaced by T.
So inclusion, c.122A>T means in the coding region, at position 122, the A nucleotide is replaced by the T nucleotide and results in SNP. Now this is a type of transversion SNP that occurred in the coding region.
Take another example. nc.1734G>A. Comment below and let me know, what does it show?
Now for rs321012, the ‘rs’ is the reference SNP cluster ID. This system was developed by NCBI and is universally accepted. This is an ID given to a particular SNP.
Read more: What is Copy Number Variation and How to Detect it?
Applications of SNPs in Genetics:
SNP is an important genetic marker for research and diagnosis. SNP markers are used in various fields including medical science and research, plant research, agriculture, bacteriology, microbiology and environmental studies.
The list of SNP marker applications is too long. So it is not possible to cover everything. I have enlisted some important applications here, comprehensively.
- SNP marker is popularly used in population genetic studies. Scientist studies SNPs from the population and identifies common, rare, pathogenic and non-pathogenic SNP. Thereby they can understand the genetic composition of the population and susceptibility for a particular disease.
- Next, it is widely used in medical research and diagnosis. Scientists identify SNPs and try to link them with a disease, condition or trait. By doing so, they can understand the likelihood of disease occurrence.
- Various SNP markers are now directly used in disease diagnosis. For example, a single SNP is used for the diagnosis of sickle cell anemia.
- It is also used in linkage disequilibrium studies.
- In recent times, it has been widely used in genome-wide association studies. Millions of SNPs have been analyzed and used to draw connections between SNPs and diseases, traits, phenotypes or conditions.
- SNP studies are employed in the personalized medicine research field as well. Here an individual’s SNP map has been prepared against the effect of a particular drug or a group of drugs. Researchers can evaluate the effect of a drug at a genetic level.
- Such studies help develop personalized treatment and medication options for patients.
- SNPs are also an important marker for evolutionary studies. By tracking back the SNP evolution, scientists can understand the process of evolution and the occurrence of genetic alterations.
- As aforementioned, SNPs are also used in agriculture, plant breeding, plant research, biomedical research, phylogenetic and taxonomic studies, animal studies, screening programs and pre-natal studies.
SNP databases:
As the SNP is an important marker in genetic research, various databases are developed and maintained to submit and use SNPs and conduct genetic research. Here are a few known and important databases.
| Database name | Description | Link |
| dbSNP | Developed by the National Centre for Biotechnology Information (NCBI). A comprehensive SNP data. | https://www.ncbi.nlm.nih.gov/snp/ |
| ClinVar | Developed by NCBI. It’s a database of clinical SNP variances that are associated with a disease or medical condition. | https://www.ncbi.nlm.nih.gov/clinvar/ |
| HapMap | Provides SNP data and linkage disequilibrium information. | https://www.genome.gov/10001688/international-hapmap-project |
| SNPedia | It’s a community-curated resource. | https://www.snpedia.com/ |
Wrapping up:
Single Nucleotide Polymorphism is a valuable genetic marker, particularly for population studies. Techniques like GWAS help in determining the risk of the likelihood of disease occurrence by SNPs.
Recent research also suggests that SNPs are associated with psychological and behavioral traits. SNPs can further alter gene expression profiles for a gene or group of genes as well.
Moreover, such single-base alterations are lucrative markers for cancer research and diagnosis. So conclusively, it’s difficult to cover everything in a single article. We will continue this series and write more articles about the SNP and its association with various conditions.
Sources:
TM Gabor and K Ian et. al., 1999. “A general approach to single-nucleotide polymorphism discovery. Nature Genetics 23; 452-456. Link: A general approach to single-nucleotide polymorphism discovery.


