“I motif or the intercalating motif is a four-stranded cytosine rich region highly unlike the normal DNA double helix.”
Although it is commonly observed in vitro, the first time it was reported in the human cells by Zeraati M, David B. L, Scofield P and coworker in the year 2018.
Before understanding the structure of the i-motif, we have to understand some basic genetics.
Key Topics:
Key information related to the present topic:
The majority of the human genome is made up of the noncoding DNA sequences (~98%). These sequences are located on the telomeric and centromeric region that are various introns, regulatory elements, enhancers, promoters, transcriptional factors, recognition sequences, intervening sequences and other non-protein coding DNA.
The non-coding DNA sequences are particularly G and C rich repeated sequences which might be conserved since evolution.
Three types of DNA forms are observed in nature: A form DNA, B form DNA and Z form DNA.
Under the common physiological conditions, the commonest DNA form is right-handed B-DNA present in the coding regions of DNA.
However, the non-coding regions can fold beyond the B forms such as hairpin structure, G-quadruplex, Z form DNA, i-motif or unstable A-form DNA.
Highly GC rich regions often form the hairpin structure whereas the high G rich region forms the unusual and unstable G-quadruplex.
But during the last decade of the 1900s, another unusual DNA form is reported called intercalating motifs.
The intercalating motif or i-motif forms in the high cytosine rich regions of the non-coding DNA.
The DNA is made up of the three important components viz phosphate, sugar and nitrogenous bases.
The sugar in DNA is deoxy sugar (D-sugar), interestingly, the D-sugar makes DNA, a DNA and differentiate it from the RNA.
The phosphate of the DNA is triphosphate, due to the negative charges on the phosphate, the DNA is negatively charged.
Read more on DNA, nucleotide and D-sugar: DNA story: The structure and function of DNA
The nucleotide joints to one another by the phosphodiester bond on the same strand and by a hydrogen bond with another strand.
Two hydrogen bonds between adenine and thymine and three hydrogen bond between cytosine and guanine stabilize the DNA.
This covalent bonding gives DNA a double-helical structure. However, the structure of the intercalating motif is a bit different than the ds DNA.

Common names: i-motif, intercalating motif, DNA i-motif, intercalating DNA motif, hTelo i-motif etc.
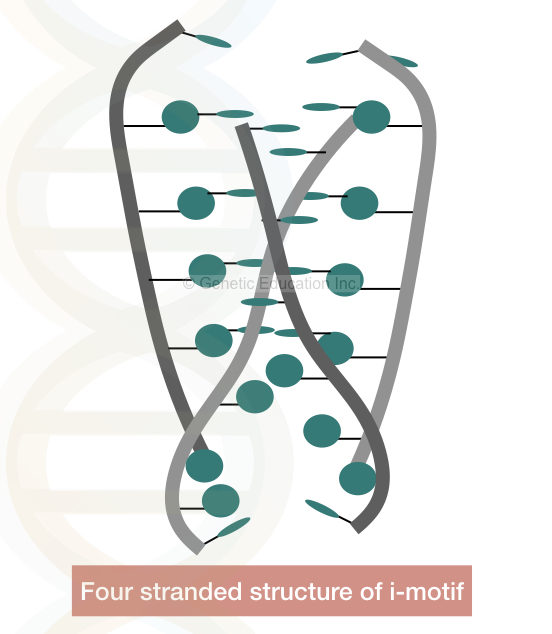
The structure of i-motif
The I motif structure is first reported in the year 1990 but the in vivo presence in the living human cell is reported recently.
The gene expression is regulated by the regulatory regions such as promoters and enhancers.
As we discussed, the regulatory regions are non-coding DNA sequences that mainly comprise the cytosine and guanine.
As the i-motif happens in the cytosine rich regions, the structure is reported more in the gene regulatory regions and therefore it affects the gene expression process.
Structurally, the i-motif is developed due to the complementary base pairing between the cytosine bases which makes a four-stranded tetramer.
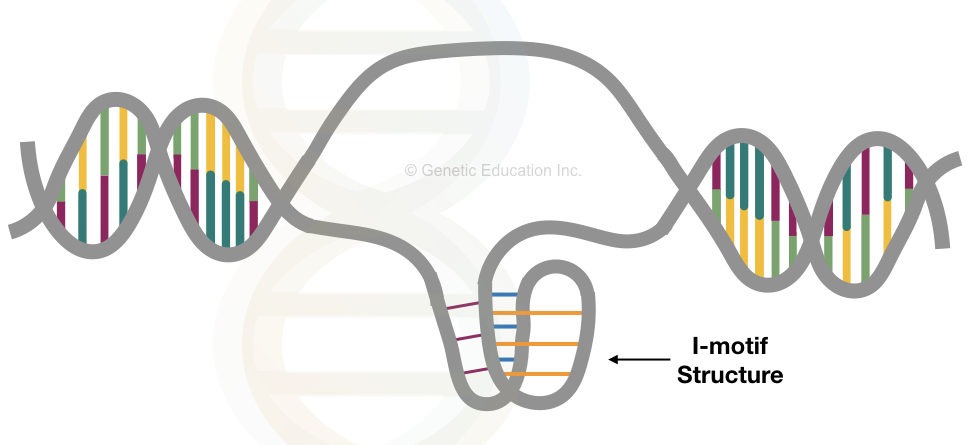
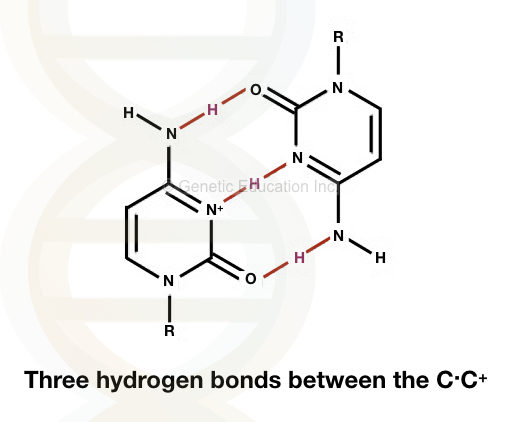
Instead of the standard Watson and Crick base pairing, the intercalating motif structure follows an unusual Hoogsteen base pairing.
It is formed by unusual base pairing between C.C+. Three hydrogen bonds between the two alternative forms of cytosine make it stronger than the standard base pairing.
The base-pairing is observed between the normal cytosine and protonated cytosine.
Structurally, the distance between the two bases in the i-motif is 3.1Å and the helical twist between the two bases are 12 to 16°.
These data indicate that the structure is similar to A DNA as well as B DNA but the structure is not fully explained by any of these models.
See the Image of base pairing between C.C+
The stability of the i-motif is depended on the pH
The stability of the structure is determined by the melting temperature of the i-DNA and the pH of the medium.
The melting temperature is a temperature at which the structure opens.
Although the i-motif structure is more stable between the pH 5.0 to 6.0, the in vivo structure of the intercalating motif is even stable at the physiological pH (at pH 7.0) and room temperature.
Nevertheless, at the room temperature and physiological pH, more than 5 cytosine molecules are required to form a stable i-motif structure.
Even in these conditions, the structure is not fully stable as it appears and disappears over time at random cytosine rich regions.
The above observation suggests that the structure of the i-motif is highly pH-dependent.
Addition to these findings, the data of Zeraati M suggests that having more than 6 consecutive cytosines provides more stability to the unusual motif structure.
[epcl_box type=”information”]More the cytosine in the DNA, more stable the i-motif structure is.[/epcl_box]
I-motif occurs regularly in hTelo, regulatory region and G1-cell cycle phase:
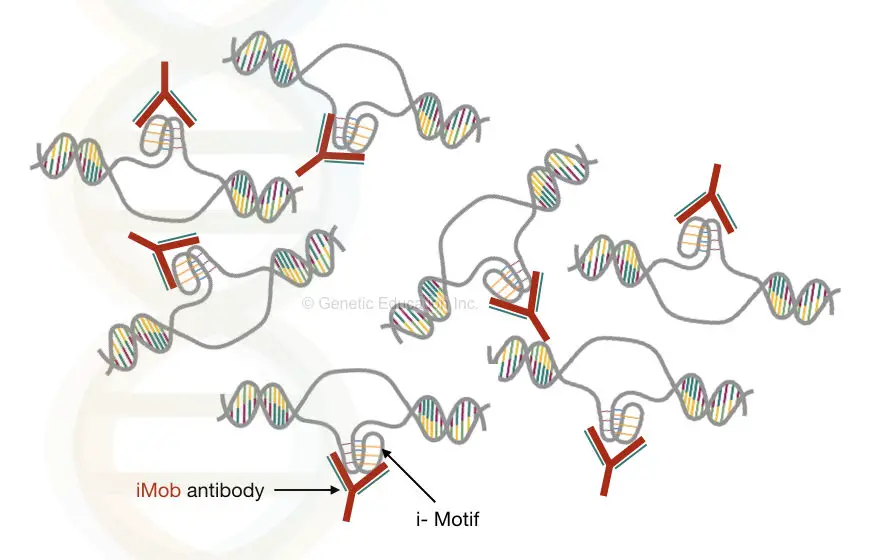
Zeraati M used the antibody fragment called “iMab” having higher binding affinity to the i-motif structure.
Using iMab, they had recognised i-motif structure at the hTelo, human telomeric regions.
The human telomeric regions are the highly repeated DNA regions majorly made up of the cytosine and Guanine rich regions.
Addition to this, the structure is also found in the transcriptional regulatory regions of a DNA.
Interestingly, the i-motif is also observed during the G-phase of the cell cycle.
Collectively, We can say that the i-motif is involved in cell growth and reproduction helps in recognition of transcriptional factors and regulation of gene expression.
The iMab (Anti-i-motif) antibody:
The iMab antibody is a monoclonal antibody designed from the scFv (Garvan-2 human single-chain) cell library through the phage display.
The antibody is first used by Zeraati et al., 2018 for the identification of i-motif structure in vivo.
Interestingly they found that the iMab has a higher affinity to the i-motif but it can not bind to the intercalating motif similar structure like G-quadruplex, hairpin DNA, miRNA or structural mutation on the telomeric regions (reported by the Zeraati et al.).
The antibody is able to detect any of the i-motif structure located on centromeres, promoters, regulator, proto-oncogene or telomeric region of the DNA.
Protonated cytosine responsible for this unusual structure:
The protonation is a process in which the proton (H+) added to the molecule or the atom which forms the conjugate acid.
[epcl_box type=”notice”]The i-motif structure is more stable in the acidic conditions.[/epcl_box]
For the formation of the C.C+ structure, one of the cystine bases must become protonated.
The structure is made up of the two parallel double strands with more than 6 cytosines having two antiparallel duplexes.
See the image below,
Due to the protonated cytosine, the negative charges of the phosphate back reduces which stabilises the DNA even in the physiological conditions.
At decreased pH the structure cannot forms due to the protonation of all the cytosine bases while at increases pH the structure unfolds.
As we earlier discussed, the structure of the motif is dependent on the pH changes hence as the physiological condition of the cell changes, the structure forms and deforms.
Scientists are believed that the i-motif structure is formed due to the DNA supercoiling. The DNA of us are negatively supercoiled which induces the formation of this unusual structure.
Zeraati M et al, concluded that “the I motif is formed in the promoter and telomeric regulatory regions of the genome hence play an important role in the genome regulation.”
Fewer published data are available which supports the role of the i-motif in human genome regulation, still, the discovery of the i-motif will add a new dimension to the genomic research in forthcoming years.
Original article: Nat Chem. 2018 Jun;10(6):631-637. doi: 10.1038/s41557-018-0046-3. Epub 2018 Apr 23.
Read some interesting articles:
- 20 Incredible DNA Facts: The 20th One Is Actually Shocking
- PCR reaction: Ten secrets that nobody tells you
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.