DNA probes are single-stranded and labeled oligonucleotide sequences used in FISH, PCR and DNA sequencing. Learn about DNA probes, types, examples and various types of labeling techniques. Also, explore applications of DNA probes, in this article.”
Genetic techniques help study genes, diseases and related mechanisms. Common genetic testing techniques that we use routinely in our lab are FISH, karyotyping, PCR, DNA microarray, DNA sequencing and restriction digestion.
Among these techniques, FISH, PCR, DNA sequencing and microarray have one common special requirement— DNA probes. It’s like a detector that helps decode the mystery of the experiment.
Without a DNA probe, we can not conduct such experiments. DNA probes carry the information which we use to study a target sequence or gene. and thus, have crucial importance.
So in this article, we will learn about DNA probes and their significance in genetic research. We will also shed light on its types, examples and labeling process.
If you are a newbie student, a budding scientist or just a curious molecular biologist, this article will help you to decode the mystery of DNA probes. You will surely learn one of the significant aspects of molecular biology in this article.
Stay tuned with this comprehensive guide.
Related article: Differences Between Probe Vs Primer.
Key Topics:
What are DNA probes?
DNA probes are short, single-stranded and labeled complementary oligonucleotide sequences used to hybridize with the target gene or sequence.
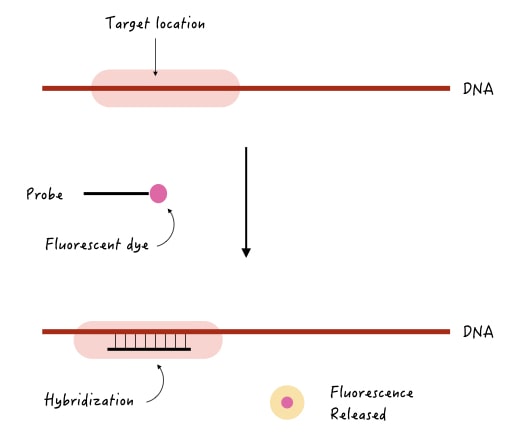
Let’s take an example to understand it.
Suppose we wish to know if a gene is present in the sample or not or does it contain some deletion or mutation. A probe is used to detect our target gene should have three properties,
- It should be complementary to the target gene.
- It should be single-stranded.
- It should have a label or a detectable tag.
Now, when we use this probe in the experiment, it will find a complementary region on the target gene, if present; hybridize there. The label tag will release and emit a detectable signal.
A detector records the signal, for example, let’s say we have used a fluorescent tag, then fluorescence will be released. After detection, the computer program will record the signal as a positive hybridization.
If the complementary region is not present at the target location, the probe will not bind and not give signals. Check out the image below to understand the concept.
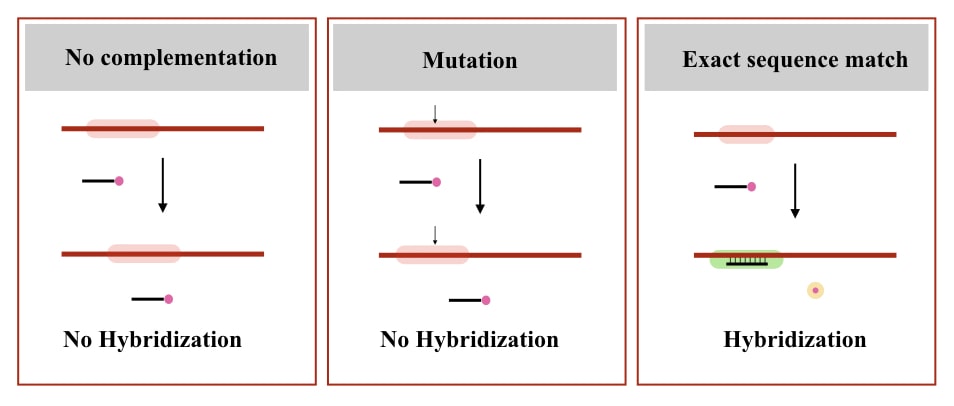
So here are the properties of a DNA probe (now some technical notes):
- DNA probes are artificially synthesized oligonucleotide sequences.
- It should be Single-stranded.
- Consist of 20 to 1000 nucleotides, depending upon the requirement of the experiment.
- Should contain a detectable tag by labeling.
- Should have complementation with the target sequence.
- Should have less GC content.
- Should have less hairpin formation capacity.
- Should not hybridize within.
Note: some FISH probes are larger in length and range from 1 to 10Kb.
Note that DNA probes are just like DNA primers, but are a bit longer and contain a detectable tag or label. Ideally, DNA probes can be labeled with fluorescent, radioactive, Biotin, chemiluminescent, Enzyme or any chemical labeling depending on the requirement of the experiment.
We will learn about each type of DNA probe labeling technique in the upcoming section, but before that let us see different types of ‘DNA probes.’
Types of DNA probes:
Based on the application, various types of DNA probes are Hybridization probes, PCR probes, Microarray probes (cDNA probes and oligo probes), Random probes and aptamers.
Hybridization probes:
Hybridization probes are single-stranded oligo-sequences used in hybridization-based detection techniques like— FISH, in situ hybridization and microarray.
FISH probes:
FISH (Fluorescence in situ hybridization) probes are single-stranded and labeled DNA probes that directly bind on chromosomes. FISH probes are a bit longer than the conventional probe.
The FISH probe length also depends on the requirement of the assay; for example, general FISH probes are a few hundred nucleotides long. While special types of probes for larger chromosomal alteration are a few thousand nucleotides long.
FISH probes are used for the detection of larger deletion, duplication and translocation which can not be assessed by conventional karyotyping.
- Locus-specific probe:
Locus-specific probes are designed and used to study a specific gene or locus on the chromosome. Thus, used in gene mapping. It can also detect copy number variation within the target gene or locus.
Notedly, multi-locus probes are used to detect multiple loci simultaneously from the sample on the same or different chromosomes.
- Alphoid probes:
Aphoid probes are specially designed repeat sequence probes, used to hybridize with the highly repetitive sequences present on the telomere and centromere. Thus, also known as telomeric, centromeric or repeat sequence probes.
Particularly the centromeric-Alphoid probes have been employed in the detection of numerical chromosomal abnormalities.
- Whole chromosome probes:
Whole chromosome probes often known as multi-FISH probes or multi-color FISH probes are specially designed probes that bind at different locations on various chromosomes.
It’s a mixture of different lengthened probes having different color tags. Each set of probes is derived from a single type of chromosome and binds there only. It’s used in chromosomal rearrangement studies.
The technique in which the multi-color FISH probes are employed is known as spectral karyotyping.
Microarray probes:
The microarray technique can be said to be an advanced version of the FISH. It can detect thousands of loci or chromosomal regions simultaneously using the same hybridization technique.
Much like the FISH probes, microarray probes are also oligonucleotide probes but comparatively shorter in length. One important thing about these known sequence probes that makes them unique is that they are immobilized on a solid surface.
The sample is added to it for hybridization. The DNA sample when applied on the chip, probes hybridize with their complementary regions if present in the sample and accordingly provides signals.
Microarray probes have been extensively used to study thousands to millions of known chromosomal regions in a single run.
PCR probes:
PCR probes have been widely used in quantitative gene analysis. Along with the sequence-specific primer, a sequence-specific hybridization probe is also used. So, when the amplification encounters the hybridization, probe release gives a positive amplification signal.
qPCR probes are important elements for the detection and quantification of amplification, gene and microbial load.
Note:
Molecular beacons:
Molecular beacons are hairpin-shaped, specially designed-probes for high sequence specificity. Sequence specificity is a crucial parameter in any hybridization process to achieve excellent results. Here, In the absence of the complementary region, the probe will remain in the hairpin shape.
The fluorochrome and quencher dye will remain attached but when it finds the complementation, the hairpin open-ups and simultaneously separates the fluorochrome and quencher molecular which gives a fluorescence signal.
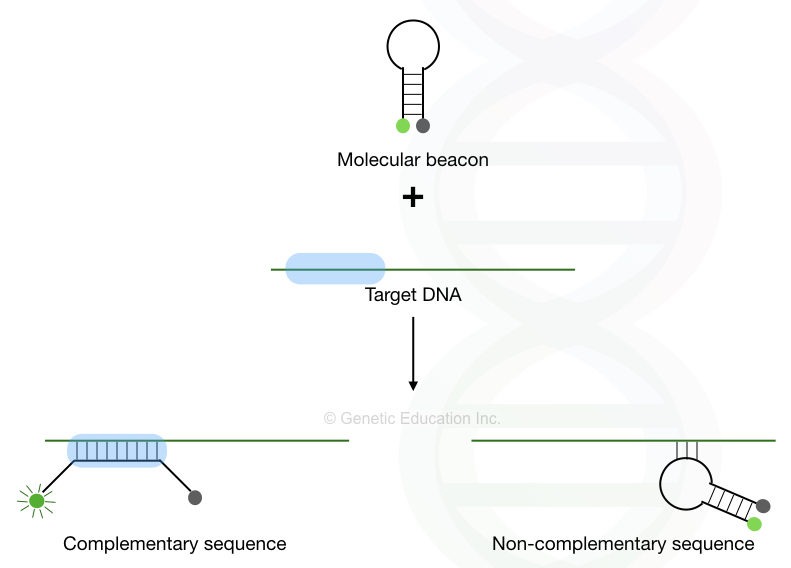
To learn more about molecular beacons, you can read this article: Structure and Function of Molecular Beacon Probes.
Aptamers:
Aptamers are DNA or RNA oligonucleotide and single-stranded probes that fold into a specific structure to bind with the target molecule like the proteins. Aptamer probes are widely used in clinical research, diagnosis and therapeutics.
Random probes:
Random sequence probes are also known as degenerative probes which are a mixture of random, known and single-nucleotide sequences. It can randomly bind to various known genomic regions.
However, it becomes a valuable tool when the sequence is entirely unknown or when multiple closely-related sequences need to be detected. It allows us to detect the presence or absence of the target sequence in our sample.
Types of Probe Labeling:
Probe labeling is a technique to incorporate a detectable tag in the nucleotide sequence. It allows us to know if the target sequence is present or not. Based on the requirements, common types of probe labeling are fluorescent, Radioactive, enzymatic, biotin and chemiluminescent labeling.
Fluorescent probe labeling:
In this technique, a fluorescent dye is attached to the 3’ end of the probe. After hybridization, the released fluorescence can be detected using fluorescence microscopy or a scanner.
Common fluorescent dyes, used for DNA probe labeling are Cy3, Cy5 and Fluorescein isothiocyanate (FITC). These probes are widely used in the FISH and related techniques.
In addition, one amazing application of fluorescent probes is that they can detect multiple sequences using multiple different colored probes in a single reaction (Multicolor FISH).
Radioactive labeling:
Radioactive labeling is the most effective and traditional way to prepare the nucleotide probes. In this technique, a radioactive isotope is used for labeling and detected using autoradiography.
Common radioactive isotopes are 32P and 35S used to prepare various types of probes, however, due to their hazardous nature, radioactive labeling isn’t recommended. Common examples of the use of radioactive labeling are the probes used in traditional blotting, sequencing and DNA fingerprinting.
Biotin labeling
Biotin labeling is the modern and safer probe labeling technique that allows highly sensitive detection and amplification by multiple binding approaches. Here, a biotin tag is attached to the probe using artificial means.
Biotinylated probes having a fluorescent dye or enzyme conjugate can be detected using streptavidin/avidin. Biotinylated probes are safe to use.
Chemiluminescent labeling
The present technique uses a chemical reaction to produce a detectable signal in the form of light emission, instead of radio emission or fluorescence. For example, a probe with an alkaline phosphatase label can undergo a chemical reaction to produce a light signal.
The signals are detected using autoradiography.
Enzymatic labeling:
In the enzymatic labeling technique, an enzyme or protein can be incorporated into the probe which when reacts with the substrate, gives a signal for hybridization. Enzymatic labeling is less advisable, as having less specificity.
Applications of DNA probes:
DNA probes have important significance in various molecular genetic techniques including DNA sequencing, PCR, FISH and microarray. Here are some of the valuable applications of DNA probes.
DNA probes are widely used in various genetic research including gene mapping, gene study, gene regulation and mutation detection.
It is also used in the diagnostic industry to detect various diseases. Genetic mutations associated with various diseases can be screened. A high-resolution FISH is used for the detection of chromosomal alterations.
Probes used in the techniques like qPCR and DNA sequencing can quantify the template and help determine the sequence, respectively.
It is also used in various genetic testing modules, DNA fingerprinting, forensic analysis, environmental studies, cancer and genetic engineering.
Limitations of DNA probes:
Probe design is a tedious, complex and costly procedure with needs expertise. Here are some of the limitations of DNA probes.
Poorly designed probes have lower specificity. It may bind to other partial complementary regions and give false signals.
Probe design and optimization is a tedious and complex process. Each probe should have to be tested for the correct length, complementation and labeling type. Poorly designed probes can cause serious analytical problems.
The sensitivity of DNA probes highly depends on the technique selected for the labeling, the type of label incorporated and the technique used for detection. Due to this reason, some probes have low sensitivity.
High sequence-specificity is also a problem in some experiments, as any mutation or sequence variation disallows probe hybridization.
One of the major limitations of nucleotide probes is that they can degrade over time and under the influence of environmental factors. Degradation directly impacts the performance, thereby the success of the experiment.
Read more: A Brief Introduction To Cytogenetics [Karyotyping, FISH and Microarray].
Wrapping up:
DNA probes are crucial for almost all molecular genetic experiments including FISH, microarray, PCR and DNA sequencing. However, the sensitivity highly depends on the labeling technique and label type used.
Fluorescent labeling is the most common, routine and safe probe labeling technique used in varieties of techniques. Commercially available probes are now ruling the market as different probes are now available for consumerized experiments.
I hope you like the present article. Please share it and bookmark our page.


Pingback: Labeling of Probes- Overview - Biology Ease