“Gene expression microarray is a laboratory genetic technique used to determine the expression of many genes simultaneously. Explore gene expression microarray, its process, advantages, applications and limitations in this article.”
Genes are like a working manual that encodes information into functions. Each gene represents each or many functions. While the cohort of many genes regulates various pathways.
For example, the P53 gene has a pivotal role in cellular differentiation, however, the same gene regulates various biological pathways when working with other genes. Scientists are now very well aware of the function of many candidate genes in various pathways.
Now they are fascinated to study the expression of genes which has a direct impact on associated phenotypes. Gene expression is in which amount a gene produces a protein. So that matters a lot– the amount/quantity!
Fortunately, techniques like qPCR or DNA microarray allow us to extensively investigate gene expression from various pathways or tissues. There are different types of microarray-based genetic tests available. DNA or cDNA microarray often known as gene expression microarray has been used to study gene expression.
It is powerful enough to detect gene expression of many genes. In this article, I will teach you about one of the most important genetic techniques: gene expression microarray. I will explain its principle, process, applications, advantages and limitations.
At Genetic Education Inc. we re-discover knowledge and present it in a layman. This article will educate newbies as well as researchers about microarrays.
Stay tuned.

Key Topics:
What is Gene Expression microarray?
First, make it clear that gene expression microarray is a type of DNA microarray, we can say. It has other names like expression microarray, gene array, DNA chip, gene chip, expression array, DNA hybridization array or oligonucleotide expression array, cDNA microarray, etc.
All these represent a similar technique— microarray.
Also, note that the expression microarray is entirely different from the chromosomal microarray. Both techniques are employed for different purposes with the same principle and with slight modification.
In this article, we will only cover gene expression microarray and its principle. So how do I define this?
Gene expression microarray is like a microscope that looks into the tissue or biological sample and checks the number of genes that encodes proteins. So it basically gives us an idea about gene activity.
It contains a chip on which specialized ‘oligos’ specific to target genes are immobilized (we will discuss it later) and a scanner to scan the chip. We just have to apply the sample on the chip and allow it to react.
The chip contains spots for thousands to millions of genes, each representing a specific message for a specific gene. The composition of the chip depends upon the chip type and requirement of the experiment.
Let’s quickly go through some historical milestones.
In 1996, Southern and co-workers developed the first high-density oligonucleotide microarray and laid the foundation for DNA microarray technology. Southern had developed the Southern hybridization techniques. From where the idea came from.
But the original idea of hybridization was first reported in the research published by Grunstein and Hogness, 1975. They screened the randomly cloned E coli DNA using radiolabeled probes. This experiment can be considered the very first DNA hybridization experiment using labeled probes.
The same approach was adopted by Gergen et al. (1979) to prepare the very first array. However, Lehrach et al. automated the process in the early ’90s. During the late ‘90s, Schena M and Brown P developed a cDNA microarray, using mRNA reverse transcription, and performed gene expression studies, independently.
From 1994 to 96, Fodor S et al. developed the first commercial DNA microarray platform at Affymetrix. Interestingly, the first microarray was developed to detect mutations from the HIV-1 reverse transcriptase and protease gene. Although, the initial technique was entirely different from the present-day technique which utilized photolithography to synthesize probes on a solid surface.
The modern-day DNA microarray first appeared in 1996 when Derisi et al. prepared a high-density spotted DNA array on a glass slide.
The gene expression cDNA microarray was first used in cancer studies by Golub T et al. (1999). Afterward, the microarray market started booming, and Affymetrix emerged as a pioneer in microarray technologies.
Afterward, from the late 2000s microarray technology started penetrating into various fields including diagnosis. Now, cancer research, gene screening, gene expression studies and transcriptomics studies are highly reliant on DNA microarrays.
Now coming back to our gene expression microarray.
In the expression microarray we need to use the mRNA and hence, isolate mRNA in the very first step. The reason behind that is here.
Replication copies the entire genomic content of a cell, as it is! Afterward, it transcribes into the mRNA. mRNA is a final transcript manufactured from the coding regions of the DNA. And translate a protein in the final step. Various mRNA transcripts are formed for various proteins depending on the requirement of the cell.
This means that the amount of mRNA is directly related to the amount of protein formed which is the amount of gene expression. A gene can be expressed when the mRNA translates into a protein.
That’s the reason, we isolate mRNA, and convert it into cDNA for microarray.
Important terminologies:
| Terminology | Explanation |
| Probes | Probes are short, single-stranded and labeled complementary sequences of nucleotides (DNA). |
| Hybridization | Process of binding complementary single-stranded DNA or RNA sequences. |
| Reverse transcription | Process of converting the mRNA into cDNA. |
Principle of Microarray:
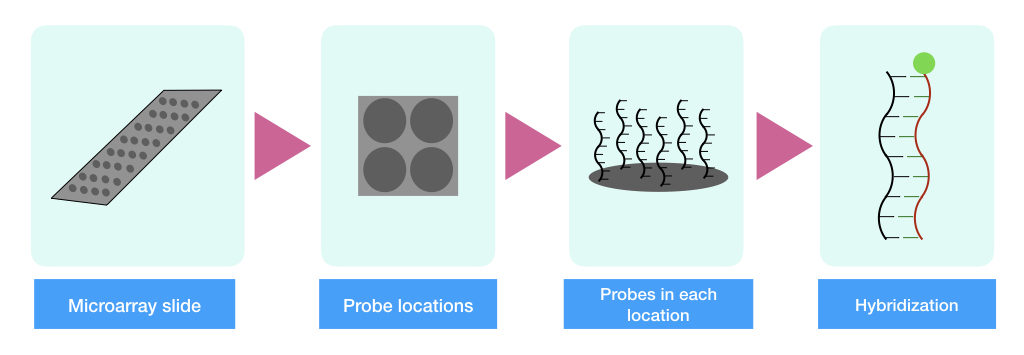
The principle of microarray relies on the hybridization between template DNA and probes. The short and single-stranded probes are immobilized on a solid microarray chip whereas the sample mRNA is first converted into the cDNA, fragmented, labeled and allowed to hybridize with the probes.
The complementation between the probes and the target cDNA includes hybridization (bound) and releases fluorescence signals. The microarray scanner scans the slide, collects the fluorescence signal and measures the intensity. Higher the fluorescence emission, the higher the level of gene expression.
This is a simple principle of gene expression microarray. Now let’s dive into each step and understand the entire process keenly.
Steps in microarray:
- Sample collection
- mRNA isolation
- Reverse transcription
- Fragmentation
- Fragment labeling
- Hybridization
- Washing
- Detection
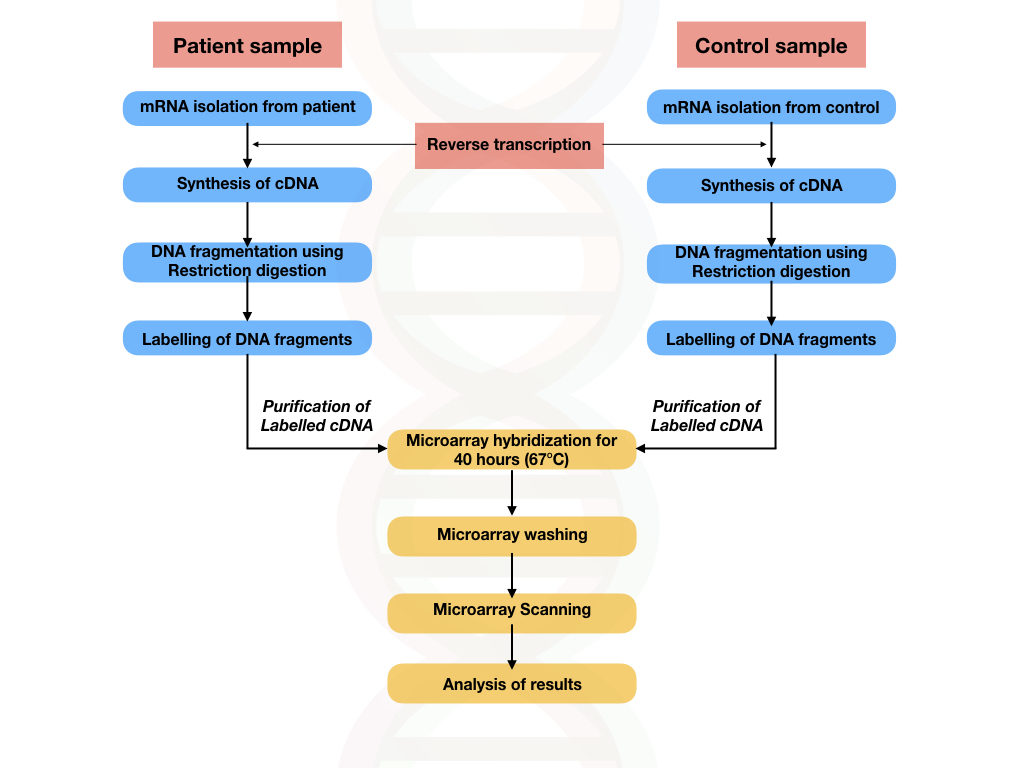
Procedure:
- Sample collection:
Any biological sample can be employed for microarray analysis. Common samples are blood, cheek swab, and solid tissues that can be used for gene expression analysis. A sample is collected and maintained using standard sample collection and maintenance procedures.
- mRNA isolation:
The next is the nucleic acid isolation step. Here, for gene expression microarray, mRNA is isolated. mRNA isolation is a sensitive procedure as the RNA is highly prone to degradation. Care must be taken and standard RNA isolation guidelines are followed during isolation.
From the total RNA, only mRNA can be isolated using the specialized Poly- d(T) containing columns. The mRNA contains a long poly-A tail which binds with the poly- d(T) column and extracts.
Later on, the extracted mRNA is purified, quantified and stored (if required) using the standard mRNA storing procedure. To learn more about how to isolate mRNA from the total cellular RNA click the link and read the article.
- Reverse transcription:
Reverse transcription is a process in which the mRNA is converted into cDNA. The reverse transcription is performed using reverse transcription PCR and reverse transcriptase enzyme.
RT-PCR can generate copies of cDNA from the target mRNA. Later, the cDNA is sent for fragmentation and labeling.
- Fragmentation:
In this step, the cDNA is fragmented into a few Kb long fragments using artificial fragmentation techniques. Restriction digestion is one such technique of DNA fragmentation.
Related articles: DNA fragmentation- Techniques, Importance and Applications.
- Fragment labeling:
Fragment labeling is a crucial scheme in this entire procedure, as the success and precision of results depend on it. In this step, a detectable tag is incorporated with each fragment to make it detectable.
Usually, in modern-day molecular biology, fluorochromes are commonly used as a labeling tag. It can be incorporated at the end of each fragment. When hybridization occurs, tags are detached and released fluorescence signals.
Two common types of fluorescent dye used in the DNA microarray are Cyt3 and Cyt5. Here, Cyt3 is a blue dye and is used to label the sample DNA fragments whereas Cyt5 is a red dye and is used to label the control DNA fragments.
A single washing step is performed to remove the unbounded dyes from the samples and allow for hybridization.
- Hybridization:
The control and patient samples (which we have prepared) are applied to the microarray chip. Each fragment hybridizes with its complementary probes, pre-immobilized on the chip.
The bound fragments release fluorescence, for example, if the patient’s DNA fragment will hybridize, the intense blue signal will detect and vice versa for a normal sample.
Two or three washing steps have been performed to remove the unbound fragments. Washing increases detection resolution by removing unwanted signals or background noise. Note that many washing steps may decrease the hybridization efficiency.
- Microarray scanning and Detection:
The microarray scanner is an important part of the entire system as it detects the signals. Hybridization releases the tag and so the fluorescence. A microarray scanner is a specialized fluorescence detector that can record every fluorescent signal emitted from the chip.
The scanner scans the entire chip many times and recovers every possible signal and sends the data to the software.
- Data analysis:
A special computer software collects, processes and analyzes the information generated by the scanner and starts analyzing the data. First, it will start categorizing the signals into normal and patient signals depending on the pre-installed software information.
The scanner measures the intensity of the signal and sends it to the computer software which converts it into numerical values. The program extracts the intensity value from each spot and groups it accordingly.
The amount of fluorescence emission is directly proportional to the amount of hybridization which means the amount of DNA fragments bound to the probes.
Thus,
“The more the hybridization, the more intense the signal and the more gene expression.”
Advantages:
Microarray is amazing! It enables scientists to study thousands of DNA sequences using pre-existing knowledge. Here are some of the important advantages.
Quantitative analysis:
A gene expression microarray gives quantitative data on gene expression for thousands of genes or the entire transcriptome of a tissue or cell type. This can help us to understand which gene expresses in which amount.
High throughput:
One of the important advantages of DNA microarray is its high throughput. Meaning, it can screen thousands of genes, millions of alterations and polymorphisms from the sample and give us quantitative data.
Speedy:
It’s realistically impossible to study the expression of thousands of genes, one by one. The present assay can do this job in a single run and thus, save a huge amount of time. A single experiment can be completed in 2 to 3 days starting from sample preparation to reporting.
It’s insanely fast!
Easy to perform:
The microarray process is very simple and includes no lengthy, tedious and complex sample processing pre-preparation steps. Labeling, hybridization and washing steps are so simple to perform.
Customization:
DNA microarray is highly customizable. Microarray chips are available depending on the requirement of the experiment. A Genome-wide chip, single gene chip, disease-associated genes chip, etc are now available.
For example, a genome-wide microarray chip can study the entire genomic variations or gene expression while a dedicated breast cancer gene panel chip covers only the alterations associated with breast cancer or the BRCA1 and BRCA2 gene.
Easy analysis:
No advanced computer knowledge or software is required to study and interpret the microarray results. An in-build microarray software system is powerful enough to study the results.
Genome-wide coverage:
The microarray has the potential to study the entire transcriptome of a cell. Meaning has entire genome-wide coverage.
Sensitive:
Expression microarray is a highly sensitive technique that gives us very precise and accurate data.
Disadvantages:
Limited dynamic range:
The microarray can study thousands of targets whether genes, alterations, SNP or indels but can give biased results. Technically, I have noticed from my personal experience that it can’t interpret very high and very low gene expression signals, precisely.
For example, It has poor sensitivity for very low gene expression and high saturation for very high gene expression. This creates a bias in results and the gene expression profile. However, for the majority of expression studies, it provides adequately accurate results.
Costly:
Any microarray technology is insanely costly. In addition, customized microarrays are even costlier than the standard ones. Microarray designing is a tedious process and includes steps like probe design, computational analysis, probe synthesis, chip design and array software design.
Each process needs huge investment and thus, the overall technique is a high-cost game. Nonetheless, what data and information it provides is so useful.
Non-specific binding:
Yet another major limitation of the hybridization-based microarray technique is non-specific binding and cross-hybridization. Probes can hybridize to a non-target location having high to low sequence similarities, repeat nucleotides and give false results.
Non-specific binding is a key reason for false-positive results in the microarray.
Sequence information:
The biggest limitation of any microarray is that it needs sequence information to prepare and design probes. Hence, it can only report alterations or gene expression for known genes and genomic regions.
It can’t identify or study unknown and novel alterations or genes.
Inefficient coverage for repetitive sequences:
Repetitive regions are the major hurdles in any genetic analysis including microarray. Genes or sequences with high repeat DNA have higher non-specific hybridization rate and thus, makes the assay inefficient to use.
Technical Limitations:
Any microarray has many technical limitations. It can’t study novel gene expression, can’t cover repeat and unknown genomic regions, and very high and low gene expressions. In addition, probe design, synthesis and labeling all are time-consuming and tedious prerequisites.
Applications:
Functional genomic studies:
Gene expression microarray is a pivotal technique in functional genomic studies. It helps to study how various genes regulate various biological processes and pathways.
Gene expression and epigenetic studies:
Tissue, cell or organ-specific expression of various genes can be determined and used to perform comparative studies. Gene expression microarray, thereby allowing researchers to study epigenetics. Disease or pathway-associated genes’ expression can be determined.
Disease research and diagnosis:
Microarray, nowadays, is extensively applied for disease research and diagnosis to study the gene expression profile of various diseases like cancer, neurodegenerative disease, cardiovascular disease, developmental disease, and complex polygenic disorders.
It can even study infectious diseases as well.
Toxicogenomics:
Another fascinating application of microarray is in toxicogenomic studies. Here, the effect of toxins, pollutants, mutagens and other environmental factors on gene expression patterns can be studied.
Hence, it is employed to investigate how various toxins influence pathways and associated genes.
Biomarker research and discoveries:
Expression microarray is widely used in large-scale biomarker studies and discoveries. Altered gene expression profiles for individuals or groups of genes can be established as a biomarker for pathway studies, disease diagnosis, and prognosis studies.
TFB sites:
Microarray is also used in combination with other techniques as well. For example, a microarray with Chromatin Immunoprecipitation is used to study the transcriptional factor binding site studies.
The TFB site on DNA is identified, separated and allowed to hybridize on the microarray chip. Such techniques allow researchers to investigate potential binding sites for a transcriptional factor on a gene or various genes.
Developmental biology:
Yet another crucial application of microarray is in developmental and pre-implantation studies. Scientists can now study genes associated with developmental processes, pathways and cellular differentiation.
They can determine the gene expression profile and find out the association of alteration of gene expression with various developmental problems including developmental abnormalities & delay, motor problems and neurological problems.
It is also widely used in pre-implantation genetic studies to investigate the transcriptomics of a growing embryo. Thus, it helps in IVF and other methods.
Personalized medicines:
Healthcare professionals are using gene expression microarrays for personalized medicines. Patient-specific gene expression profile or transcriptomic analysis is used to know drug resistance, treatment and therapeutic outcomes. Such studies are useful for preparing more personalized treatments for patients.
Conclusion:
I used a microarray during my PhD. It blew my mind– how a single experiment can give us such a huge amount of data! That’s the reason, it is getting recognition timely. Now, it has become a valuable and important tool in disease diagnosis.
Microarray is a simple and easy-to-perform technique and does not require an extensive technical background. It revolutionized the field of genetic analysis. Its ability to study thousands to millions of queries makes it a unique and crucial technology for future research.
I hope you like this article. Please comment, bookmark the page and subscribe to our blog.
Subscribe to our weekly newsletter for the latest blogs, articles and updates, and never miss the latest product or an exclusive offer.



