“Non-specific amplification is one of the most common problems in any PCR reaction. In this guide, I will explain the reasons and possible (technical) solutions to overcome non-specific amplification.”
Imagine, you are observing a gel, curious about your PCR results and seeing many bands instead of the desired fragment you want! Confused? Don’t know what it is and how it occurred. You may have many more questions too.
That’s technically a non-specific amplification in which you see many DNA amplification bands instead of the target one. PCR- A polymerase Chain reaction is an amplification technique that can generate copies of DNA, and is widely used in molecular genetic studies and other fields of science.
It’s a simple enzyme-governed thermocycling reaction that uses dNTPs, Taq DNA polymerase, PCR buffer, primers, template DNA and water as a reaction mixture. At a specific annealing temperature, the amplification occurs.
However, despite having lucrative advantages of PCR that make it a superior technique in molecular genetics, it’s challenging. It does not always yield the expected results. Non-specific amplification is one such issue in the PCR. It leads to incorrect, ambiguous and unwanted results.
In this article, I will comprehensively explain what non-specific amplification is, how it occurs and some of the common and technical healthy practices for troubleshooting. At Genetic Education Inc. oftentimes, we re-discover the knowledge.
However, we also write technical articles and blogs using our years of experience in the field of genetics. This article will surely strengthen your knowledge and technical skills in PCR and optimization.
Stay tuned.
Download our ebook: DNA Extraction to PCR.
Key Topics:
What is non-specific amplification?
It simply means amplification which isn’t specific. Comprehensive understanding suggests that it may create two different conditions- non-specific amplification along with target amplicon and non-specific amplification without target amplicon.
The technical definition of non-specific amplification suggests that “when the PCR amplifies unintended or random DNA sequences with or without our target sequence and produces multiple or single amplicons (or bands) of an incorrect size, is referred to as non-specific amplification.”
There aren’t more dedicated technical specifications regarding the non-specific amplification in the literature. So I have divided it into two parts.
- Non-specific amplification with multiple amplicons (or bands): in which various unwanted fragments of incorrect size are generated during the PCR reaction.
- Non-specific amplification with a single amplicon: in which a single incorrect-sized amplicon is generated during the PCR amplification.
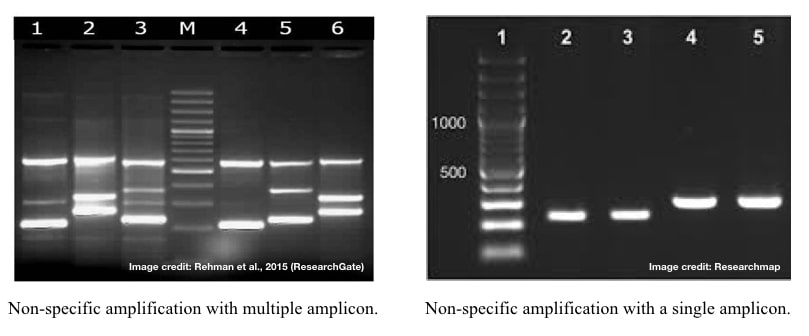
Both scenarios are classified into “non-specific amplification.” Now let’s see how it occurs.
How does non-specific amplification occur?
The non-specific amplification occurs when the annealing temperature, primer design criteria and PCR conditions are compromised with other relevant targets in the sample. When the annealing temperature is decreased, primers share sequence homology with other regions and the reaction gets contaminated, Taq DNA polymerase amplifies regions in the sample other than our target. And cause non-specific amplification.
Causes of non-specific amplification:
In this section, I will explain common causes for the present condition. We will list from the most common and high-impact factors to the least important factors.
Annealing temperature:
The most common cause for the non-specific PCR amplification is annealing temperature. When it is compromised, evident non-specific genomic amplification will occur. It usually happens at a lower annealing temperature than the desired one.
The annealing temperature is the temperature at which the primers bind to their exact complementary location on the genome. However, lowering the annealing temperature will give flexibility to primers, binding at random locations.
At very flexible annealing temperatures, even a few base pair complementation between primers and templates produce undesired PCR products. So as the annealing temperature reduces, the chances of non-specific amplification increase.
To read more, please refer to this article: What is annealing temperature and how to set and calculate it?
Poor primer design:
Another common cause for non-specific amplification is poor primer design strategies. The set of a primer used in any PCR reaction should be highly specific and complementary to the target region. The complementation should be strong enough that primers should avoid binding with other partial complement or non-complement regions.
If the primer binds with the target location but isn’t specific enough to that particular ‘target’, it binds on other genomic regions and gives multiple amplicons. To ensure the specificity and efficacy of primers, these criteria should be fulfilled.
- Design primers using software, like- Primer3.
- Use 18 to 22 nt long primer.
- The primer should avoid complementation with other genomic regions.
- The primer should avoid hairpin formation.
- The primer should have a specific annealing temperature that can’t give non-specific amplification.
Primer design isn’t that difficult, it’s a software-based process you can easily learn and perform. I have given the demo in this article: How to design PCR primers.
Primer concentration:
The least learned yet important factor that impacts the specificity of PCR is primer concentration. An appropriate concentration of primers or as advised by the manufacturer should be used in the reaction. Higher or lower concentrations will negatively impact the reaction.
A higher primer concentration means more primers in the reaction (more unwanted number of primers). So what happens, during the transition of temperature between one to another stage, or any phase of the reaction when the annealing temperature is not maintained, such unused primers bind randomly to genomic fragments and produce random and short fragments.
The ideal concentration of primers is given in the troubleshooting part of this article.
Template concentration:
Template concentration is yet another crucial factor in a PCR reaction that will surely negatively impact the reaction. At higher template concentrations, the chances of non-specific amplification increase greatly.
Follow a standard template addition protocol or take advice from an expert. The ideal template concentration in the PCR reaction is 10 to 100 ng per reaction.
Related article: What are the properties of PCR (template) DNA?
Contamination:
Non-specific amplification is also evidence of potential reaction contamination too. If any of the PCR reagents are contaminated with other sources of DNA, it may amplify multiple regions and produce non-specific amplification.
To avoid contamination use high-quality reagents and prepare reactions in a sterile and clean room or area. Keep in mind that contamination may occur at any stage between DNA extraction to PCR run. So care must be taken during each stage.
Longer PCR run:
More the PCR cycles, the more the number of amplicons and the better the resolution of bands. However, more PCR cycles are not always in favor. As the number of PCR cycles increases, it also elevates the chances of non-specific amplification too. Primers will bind to more random locations and create multiple unwanted amplifications.
A good number of PCR cycles are 30 to 35. However, to make your reaction more specific, I strongly recommend optimizing the number of PCR cycles starting from 25 cycles to 35. It saves time and increases reaction specificity.
Related article: Optimizing PCR Cycles to Get Good Results.
Primer-dimer:
Dimers are yet another common problem every PCR reaction usually faces. Partial-primer specificity sometimes produces amplicons of shorter size. Such amplicons are clustered between the region of 50 to 100 bp.
Use the appropriate concentration of primers to avoid primer non-specificity and dimer formation. The ideal concentration of primers in any PCR reaction is 10 pM.
PCR buffer:
The PCR buffer greatly increases the reaction specificity and amplification capacity. However, oftentimes, the benefits become a curse for the reaction. For example, using a higher concentration of MgCl2 will lead to non-specific binding.
The role of MgCl2 is to boost the activity of Taq DNA polymerase. MgCl2 provides a cofactor for Taq DNA polymerase to execute its catalytic activity. So at higher MgCl2 concentrations, the activity of Taq DNA pol increases unwantedly and produces non-specific amplification.
However, it’s advisable to use MgCl2 when required. Take an expert’s advice on when to use any PCR enhancers. The ideal MgCl2 concentration in a PCR reaction is ~1.5 to 2.5 Mm.
Troubleshooting for non-specific amplification:
- Use Primer 3 to design primers and set the annealing temperature for the reaction.
- The ideal annealing temperature for any PCR reaction should be between 55°C to 65°C.
- Use 10 pM primers in the reaction.
- Prepare the reaction in a sterile, clean and separate area. Follow general lab SOP for DNA extraction, purification, reaction preparation and PCR run to avoid any contamination.
- Use 10 to 100 ng DNA per reaction. Note that the purity of DNA should be near ~1.8.
- Use good practices to avoid primer-dimer formation.
- Apply 25 to 35 cycles for the PCR run. Avoid unnecessary longer PCR runs.
- Use PCR buffer or enhancers only if required. Use MgCl2 for hard-to-amplify templates only. Use 1.5 to 2.5 mM MgCl2 only.
- Use PCR controls.
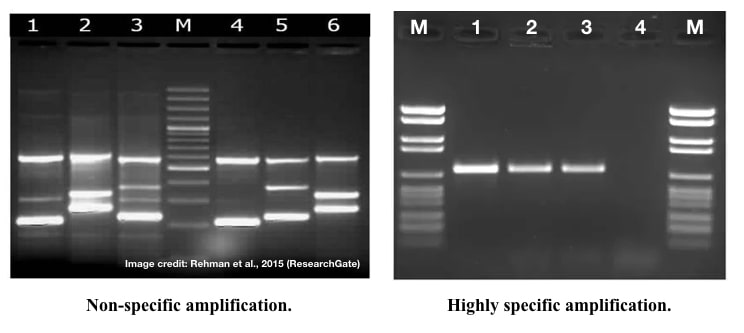
Strategies for troubleshooting non-specific amplification:
Two common strategies for troubleshooting non-specific amplification are gradient and in silico PCR.
Gradient PCR:
Perform gradient PCR to optimize the number of PCR cycles, the concentration of primers, template DNA, MgCl2 and annealing temperature. Set different temperature gradients for each parameter and evaluate results on a gel. Select the best combination by looking into the gel.
We have covered an amazing article on how to perform a gradient PCR. You can read it here.
In silico PCR:
In silico, PCR is a great way to predict the results of any PCR reaction. It’s an online tool that requires information on the primer sequence and annealing temperature and accordingly, it performs an in silico PCR reaction and gives us an idea about the possible amplification site for the particular set of primers.
To learn more read this article: In silico PCR.
Use PCR controls:
PCR controls have great value in validating PCR reactions. Use positive, negative and internal PCR controls for knowing the reaction specificity, assessing contamination and understanding the amplification conditions, respectively.
Read this article to learn how to use PCR controls: What are PCR controls and how to use them?
Wrapping up:
In conclusion, non-specific amplification in a PCR can be caused by various factors including non-specific primers, contamination, high template concentration, too many PCR cycles, too much MgCl2 and low annealing temperature.
For screening and diagnosis purposes it’s important to optimize the reaction for getting the best results using in silico PCR and gradient PCR. So this is it for the PCR troubleshooting 101 class. I hope you like this article.